Please be patient as the page loads
|
ELL_MOUSE
|
||||||
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ELL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.955643 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
ELL_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
ELL2_HUMAN
|
||||||
| θ value | 2.15048e-102 (rank : 3) | NC score | 0.924470 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
ELL3_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 4) | NC score | 0.764498 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80VR2 | Gene names | Ell3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
ELL3_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 5) | NC score | 0.769057 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HB65, Q9H634 | Gene names | ELL3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
OCLN_MOUSE
|
||||||
| θ value | 3.63628e-17 (rank : 6) | NC score | 0.568132 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61146, Q544A7 | Gene names | Ocln, Ocl | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
OCLN_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 7) | NC score | 0.571845 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16625, Q8N6K1 | Gene names | OCLN | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
TM108_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.081037 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.065754 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.034916 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ELOA2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.045413 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.044567 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.033704 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
FOXI1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.016762 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |
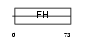
|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
CN155_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.076776 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.058607 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.041287 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CJ047_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.068171 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
FKBP5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.029295 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
GNPTA_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.036276 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q69ZN6, Q3U3K6, Q3US34 | Gene names | Gnptab, Gnpta, Kiaa1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.031899 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.028009 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
CEP57_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.065523 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.025317 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
IPR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.039553 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.055884 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
DMPK_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.005655 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
NR1D1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.013094 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.035962 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.046104 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BCL6_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.002022 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |
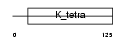
|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.059496 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.053258 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.039248 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SPA3N_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.015305 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WP6, Q62258 | Gene names | Serpina3n, Spi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3N precursor (Serpin A3N). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.024939 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
CEP57_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.062002 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.006113 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
TLE2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.014631 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
CF182_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.052391 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IYX8 | Gene names | C6orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.039707 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.025339 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.024437 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SH3G1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.018285 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62419 | Gene names | Sh3gl1, Sh3d2b | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (SH3p8). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.010599 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.047607 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FIBG_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.010444 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCM7, Q8WUR3, Q91ZP0 | Gene names | Fgg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen gamma chain precursor. | |||||
|
FKBP5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.024113 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.025752 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.013093 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.049149 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.023214 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
SOCS4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.018900 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXH5 | Gene names | SOCS4, SOCS7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
FA47A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.032994 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.043877 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.014206 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.031240 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.036731 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.054020 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.051790 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.022390 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TBC10_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.016480 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXI6, O76053, Q543A2 | Gene names | TBC1D10A, EPI64, TBC1D10 | |||
|
Domain Architecture |
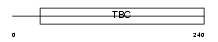
|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.021906 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.025140 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
AB1IP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.030203 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.017763 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CEND1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.011275 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
CJ006_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.020385 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.022371 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
ERCC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.020011 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92889, O00140, Q8TD83 | Gene names | ERCC4, ERCC11, XPF | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4) (DNA-repair protein complementing XP-F cells) (Xeroderma pigmentosum group F-complementing protein). | |||||
|
HBS1L_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.012093 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.030151 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.026528 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.015182 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
ZSCA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.001815 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NBB4, Q6WLH8, Q86WS8 | Gene names | ZSCAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 1. | |||||
|
BARH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.005245 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VIB5, Q66L43 | Gene names | Barhl2, Barh1, Mbh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 2 homeobox protein (Bar-class homeodomain protein MBH1) (Homeobox protein B-H1). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.025821 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.020440 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.019336 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.012431 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
FBX27_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.011371 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI29, Q96C87 | Gene names | FBXO27, FBG5, FBX27 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 27 (F-box/G-domain protein 5). | |||||
|
GLIS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.000572 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.017625 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.023912 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.027462 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.033289 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.024770 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.021850 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.020256 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
DNMBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.008623 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
ELK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.009689 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
HCN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.006697 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.029431 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LAMP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.019615 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQV4, O94781, Q8NEC8 | Gene names | LAMP3, DCLAMP, TSC403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (Protein TSC403) (CD208 antigen). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.036954 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.014817 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.022525 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.028552 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.011445 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.003589 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
FREA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.008240 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61574, Q8C5N7 | Gene names | Fkhl18, Fkh3, Freac10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein) (Transcription factor FKH-3). | |||||
|
MIS12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.014862 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CY25, Q3V261, Q5QNU7, Q5QNU8, Q5QNU9 | Gene names | Mis12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MIS12 homolog. | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.015161 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.015017 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
NFIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.019696 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70255, O09072, P70256, Q3U2I9, Q99MA3, Q99MA4, Q99MA5, Q99MA6, Q99MA7, Q99MA8, Q9R1G3 | Gene names | Nfic | |||
|
Domain Architecture |
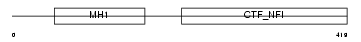
|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
PFD6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.036540 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15212 | Gene names | PFDN6, HKE2, PFD6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
PFD6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.034726 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03958 | Gene names | Pfdn6, H2-Ke2, Pfd6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
SOCS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.011284 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14543, O14509 | Gene names | SOCS3, CIS3, SSI3 | |||
|
Domain Architecture |
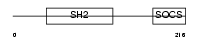
|
|||||
| Description | Suppressor of cytokine signaling 3 (SOCS-3) (Cytokine-inducible SH2 protein 3) (CIS-3) (STAT-induced STAT inhibitor 3) (SSI-3). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.007095 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.045160 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.038440 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TLE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.010646 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.018690 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
TPM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.018695 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.011116 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.061758 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
ELL_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
ELL_HUMAN
|
||||||
| NC score | 0.955643 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P55199 | Gene names | ELL, C19orf17 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
ELL2_HUMAN
|
||||||
| NC score | 0.924470 (rank : 3) | θ value | 2.15048e-102 (rank : 3) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
ELL3_HUMAN
|
||||||
| NC score | 0.769057 (rank : 4) | θ value | 2.42779e-29 (rank : 5) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9HB65, Q9H634 | Gene names | ELL3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
ELL3_MOUSE
|
||||||
| NC score | 0.764498 (rank : 5) | θ value | 1.08979e-29 (rank : 4) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80VR2 | Gene names | Ell3 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL3. | |||||
|
OCLN_HUMAN
|
||||||
| NC score | 0.571845 (rank : 6) | θ value | 4.74913e-17 (rank : 7) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16625, Q8N6K1 | Gene names | OCLN | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
OCLN_MOUSE
|
||||||
| NC score | 0.568132 (rank : 7) | θ value | 3.63628e-17 (rank : 6) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61146, Q544A7 | Gene names | Ocln, Ocl | |||
|
Domain Architecture |

|
|||||
| Description | Occludin. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.081037 (rank : 8) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.076776 (rank : 9) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CJ047_HUMAN
|
||||||
| NC score | 0.068171 (rank : 10) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86WR7, Q5W0J9, Q5W0K0, Q5W0K1, Q5W0K2, Q6PJC8, Q8N317 | Gene names | C10orf47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47. | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.065754 (rank : 11) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.065523 (rank : 12) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CEP57_HUMAN
|
||||||
| NC score | 0.062002 (rank : 13) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86XR8, Q14704, Q8IXP0, Q9BVF9 | Gene names | CEP57, KIAA0092, TSP57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin) (FGF2-interacting protein). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.061758 (rank : 14) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.059496 (rank : 15) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.058607 (rank : 16) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.055884 (rank : 17) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.054020 (rank : 18) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.053258 (rank : 19) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CF182_HUMAN
|
||||||
| NC score | 0.052391 (rank : 20) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IYX8 | Gene names | C6orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.051790 (rank : 21) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.049149 (rank : 22) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.047607 (rank : 23) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.046104 (rank : 24) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ELOA2_HUMAN
|
||||||
| NC score | 0.045413 (rank : 25) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYF1, Q9P2V9 | Gene names | TCEB3B, TCEB3L | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A2 (Elongin-A2) (EloA2) (Transcription elongation factor B polypeptide 3B). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.045160 (rank : 26) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.044567 (rank : 27) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.043877 (rank : 28) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.041287 (rank : 29) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.039707 (rank : 30) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
IPR1_MOUSE
|
||||||
| NC score | 0.039553 (rank : 31) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BVK9, Q3UCV7, Q80V00 | Gene names | Ipr1, Ifi75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intracellular pathogen resistance protein 1. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.039248 (rank : 32) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.038440 (rank : 33) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.036954 (rank : 34) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.036731 (rank : 35) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
PFD6_HUMAN
|
||||||
| NC score | 0.036540 (rank : 36) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15212 | Gene names | PFDN6, HKE2, PFD6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
GNPTA_MOUSE
|
||||||
| NC score | 0.036276 (rank : 37) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q69ZN6, Q3U3K6, Q3US34 | Gene names | Gnptab, Gnpta, Kiaa1208 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetylglucosamine-1-phosphotransferase subunits alpha/beta precursor (EC 2.7.8.17) (GlcNAc-1-phosphotransferase alpha/beta subunits) (UDP- N-acetylglucosamine-1-phosphotransferase alpha/beta subunits) (Stealth protein GNPTAB) [Contains: N-acetylglucosamine-1-phosphotransferase subunit alpha; N-acetylglucosamine-1-phosphotransferase subunit beta]. | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.035962 (rank : 38) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.034916 (rank : 39) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
PFD6_MOUSE
|
||||||
| NC score | 0.034726 (rank : 40) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03958 | Gene names | Pfdn6, H2-Ke2, Pfd6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.033704 (rank : 41) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.033289 (rank : 42) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
FA47A_HUMAN
|
||||||
| NC score | 0.032994 (rank : 43) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5JRC9, Q8TAA0 | Gene names | FAM47A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47A. | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.031899 (rank : 44) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.031240 (rank : 45) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
AB1IP_MOUSE
|
||||||
| NC score | 0.030203 (rank : 46) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8R5A3, O35329, Q8BRU0, Q99KV8 | Gene names | Apbb1ip, Prel1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 48). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.030151 (rank : 47) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.029431 (rank : 48) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
FKBP5_MOUSE
|
||||||
| NC score | 0.029295 (rank : 49) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64378 | Gene names | Fkbp5, Fkbp51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.028552 (rank : 50) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.028009 (rank : 51) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.027462 (rank : 52) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.026528 (rank : 53) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.025821 (rank : 54) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.025752 (rank : 55) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.025339 (rank : 56) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.025317 (rank : 57) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.025140 (rank : 58) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.024939 (rank : 59) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.024770 (rank : 60) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.024437 (rank : 61) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
FKBP5_HUMAN
|
||||||
| NC score | 0.024113 (rank : 62) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13451 | Gene names | FKBP5, AIG6, FKBP51 | |||
|
Domain Architecture |

|
|||||
| Description | FK506-binding protein 5 (EC 5.2.1.8) (Peptidyl-prolyl cis-trans isomerase) (PPIase) (Rotamase) (51 kDa FK506-binding protein) (FKBP- 51) (54 kDa progesterone receptor-associated immunophilin) (FKBP54) (P54) (FF1 antigen) (HSP90-binding immunophilin) (Androgen-regulated protein 6). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.023912 (rank : 63) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.023214 (rank : 64) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.022525 (rank : 65) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.022390 (rank : 66) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.022371 (rank : 67) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.021906 (rank : 68) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.021850 (rank : 69) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.020440 (rank : 70) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CJ006_MOUSE
|
||||||
| NC score | 0.020385 (rank : 71) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P9P0, Q8C041, Q8C0J5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf6 homolog. | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.020256 (rank : 72) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ERCC4_HUMAN
|
||||||
| NC score | 0.020011 (rank : 73) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92889, O00140, Q8TD83 | Gene names | ERCC4, ERCC11, XPF | |||
|
Domain Architecture |

|
|||||
| Description | DNA repair endonuclease XPF (EC 3.1.-.-) (DNA excision repair protein ERCC-4) (DNA-repair protein complementing XP-F cells) (Xeroderma pigmentosum group F-complementing protein). | |||||
|
NFIC_MOUSE
|
||||||
| NC score | 0.019696 (rank : 74) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70255, O09072, P70256, Q3U2I9, Q99MA3, Q99MA4, Q99MA5, Q99MA6, Q99MA7, Q99MA8, Q9R1G3 | Gene names | Nfic | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 C-type (Nuclear factor 1/C) (NF1-C) (NFI-C) (NF-I/C) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
LAMP3_HUMAN
|
||||||
| NC score | 0.019615 (rank : 75) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQV4, O94781, Q8NEC8 | Gene names | LAMP3, DCLAMP, TSC403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (Protein TSC403) (CD208 antigen). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.019336 (rank : 76) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
SOCS4_HUMAN
|
||||||
| NC score | 0.018900 (rank : 77) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WXH5 | Gene names | SOCS4, SOCS7 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 4 (SOCS-4) (Suppressor of cytokine signaling 7) (SOCS-7). | |||||
|
TPM3_MOUSE
|
||||||
| NC score | 0.018695 (rank : 78) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21107, Q09021, Q60606, Q80SW6, Q9EPW3 | Gene names | Tpm3, Tpm-5, Tpm5 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.018690 (rank : 79) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
SH3G1_MOUSE
|
||||||
| NC score | 0.018285 (rank : 80) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62419 | Gene names | Sh3gl1, Sh3d2b | |||
|
Domain Architecture |

|
|||||
| Description | SH3-containing GRB2-like protein 1 (Endophilin-2) (Endophilin-A2) (SH3 domain protein 2B) (SH3p8). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.017763 (rank : 81) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.017625 (rank : 82) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
FOXI1_HUMAN
|
||||||
| NC score | 0.016762 (rank : 83) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
TBC10_HUMAN
|
||||||
| NC score | 0.016480 (rank : 84) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXI6, O76053, Q543A2 | Gene names | TBC1D10A, EPI64, TBC1D10 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 10A (EBP50-PDX interactor of 64 kDa) (EPI64 protein). | |||||
|
SPA3N_MOUSE
|
||||||
| NC score | 0.015305 (rank : 85) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WP6, Q62258 | Gene names | Serpina3n, Spi2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor A3N precursor (Serpin A3N). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.015182 (rank : 86) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.015161 (rank : 87) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.015017 (rank : 88) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MIS12_MOUSE
|
||||||
| NC score | 0.014862 (rank : 89) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CY25, Q3V261, Q5QNU7, Q5QNU8, Q5QNU9 | Gene names | Mis12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein MIS12 homolog. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.014817 (rank : 90) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
TLE2_MOUSE
|
||||||
| NC score | 0.014631 (rank : 91) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVB2 | Gene names | Tle2, Grg2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2. | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.014206 (rank : 92) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NR1D1_HUMAN
|
||||||
| NC score | 0.013094 (rank : 93) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.013093 (rank : 94) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.012431 (rank : 95) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
HBS1L_HUMAN
|
||||||
| NC score | 0.012093 (rank : 96) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y450, Q5T7G3, Q8NDW9, Q9UPW3 | Gene names | HBS1L, HBS1, KIAA1038 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HBS1-like protein (ERFS). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.011445 (rank : 97) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
FBX27_HUMAN
|
||||||
| NC score | 0.011371 (rank : 98) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NI29, Q96C87 | Gene names | FBXO27, FBG5, FBX27 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 27 (F-box/G-domain protein 5). | |||||
|
SOCS3_HUMAN
|
||||||
| NC score | 0.011284 (rank : 99) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O14543, O14509 | Gene names | SOCS3, CIS3, SSI3 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of cytokine signaling 3 (SOCS-3) (Cytokine-inducible SH2 protein 3) (CIS-3) (STAT-induced STAT inhibitor 3) (SSI-3). | |||||
|
CEND1_MOUSE
|
||||||
| NC score | 0.011275 (rank : 100) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BZ05, Q80TX2, Q8C3T2, Q8VEL6 | Gene names | Centd1, Kiaa0580 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 1 (Cnt-d1). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.011116 (rank : 101) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
TLE2_HUMAN
|
||||||
| NC score | 0.010646 (rank : 102) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04725, Q8WVY0, Q9Y6S0 | Gene names | TLE2 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 2 (ESG2). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.010599 (rank : 103) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
FIBG_MOUSE
|
||||||
| NC score | 0.010444 (rank : 104) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCM7, Q8WUR3, Q91ZP0 | Gene names | Fgg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrinogen gamma chain precursor. | |||||
|
ELK4_HUMAN
|
||||||
| NC score | 0.009689 (rank : 105) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28324, P28323 | Gene names | ELK4, SAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing protein Elk-4 (Serum response factor accessory protein 1) (SAP-1). | |||||
|
DNMBP_HUMAN
|
||||||
| NC score | 0.008623 (rank : 106) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6XZF7, Q8IVY3, Q9Y2L3 | Gene names | DNMBP, KIAA1010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dynamin-binding protein (Scaffold protein Tuba). | |||||
|
FREA_MOUSE
|
||||||
| NC score | 0.008240 (rank : 107) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61574, Q8C5N7 | Gene names | Fkhl18, Fkh3, Freac10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead-related transcription factor 10 (FREAC-10) (Forkhead-like 18 protein) (Transcription factor FKH-3). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.007095 (rank : 108) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
HCN1_HUMAN
|
||||||
| NC score | 0.006697 (rank : 109) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60741 | Gene names | HCN1, BCNG1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 (Brain cyclic nucleotide-gated channel 1) (BCNG-1). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.006113 (rank : 110) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
DMPK_HUMAN
|
||||||
| NC score | 0.005655 (rank : 111) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
BARH2_MOUSE
|
||||||
| NC score | 0.005245 (rank : 112) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VIB5, Q66L43 | Gene names | Barhl2, Barh1, Mbh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BarH-like 2 homeobox protein (Bar-class homeodomain protein MBH1) (Homeobox protein B-H1). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.003589 (rank : 113) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
BCL6_MOUSE
|
||||||
| NC score | 0.002022 (rank : 114) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P41183, Q61065 | Gene names | Bcl6, Bcl-6 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 6 protein homolog. | |||||
|
ZSCA1_HUMAN
|
||||||
| NC score | 0.001815 (rank : 115) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 724 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NBB4, Q6WLH8, Q86WS8 | Gene names | ZSCAN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 1. | |||||
|
GLIS1_HUMAN
|
||||||
| NC score | 0.000572 (rank : 116) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 115 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NBF1, Q8N9V9 | Gene names | GLIS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1). | |||||