Please be patient as the page loads
|
ARVC_HUMAN
|
||||||
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARVC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995612 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.974949 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.975107 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 5.41814e-138 (rank : 5) | NC score | 0.925315 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 2.78268e-134 (rank : 6) | NC score | 0.936130 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 1.80368e-133 (rank : 7) | NC score | 0.947803 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 2.97994e-128 (rank : 8) | NC score | 0.945767 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP2_HUMAN
|
||||||
| θ value | 3.96481e-64 (rank : 9) | NC score | 0.917456 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 5.17823e-64 (rank : 10) | NC score | 0.910024 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| θ value | 8.83269e-64 (rank : 11) | NC score | 0.909052 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP1_HUMAN
|
||||||
| θ value | 1.41016e-61 (rank : 12) | NC score | 0.904457 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP1_MOUSE
|
||||||
| θ value | 1.84173e-61 (rank : 13) | NC score | 0.905882 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 14) | NC score | 0.323565 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 15) | NC score | 0.323637 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 16) | NC score | 0.405117 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
PLAK_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 17) | NC score | 0.289751 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.231515 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
APC_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 19) | NC score | 0.237598 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
PLAK_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 20) | NC score | 0.251343 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PDLI4_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 21) | NC score | 0.036198 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50479, Q96AT8, Q9BTW8, Q9Y292 | Gene names | PDLIM4, RIL | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.021922 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.116203 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
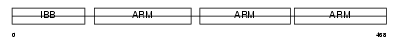
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.116274 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
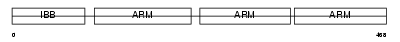
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
EDA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.034255 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.009402 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 27) | NC score | 0.018368 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 28) | NC score | 0.130150 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
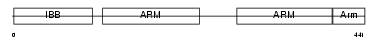
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
ARMC6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.085220 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.130195 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.025425 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ARMC6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.084574 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.072186 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.099217 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
NANO1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.034915 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WY41 | Gene names | NANOS1, NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1 (NOS-1) (EC_Rep1a). | |||||
|
XPC_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.027458 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.071548 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.037476 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
IMA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.099336 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
RTDR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.082229 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.131616 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
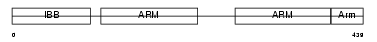
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
KLF13_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.000445 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJZ6, Q9ESX3, Q9JHF8 | Gene names | Klf13, Bteb3, Fklf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Erythroid transcription factor FKLF-2). | |||||
|
CLAP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.020678 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
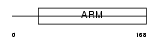
HEAT3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.074819 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
K1849_HUMAN
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.012791 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.016184 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
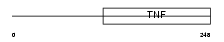
EDA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.026461 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92838, O75910, Q5JUM7, Q9UP77, Q9Y6L0, Q9Y6L1, Q9Y6L2, Q9Y6L3, Q9Y6L4 | Gene names | EDA, ED1 | |||
|
Domain Architecture |
|
|||||
| Description | Ectodysplasin-A (Ectodermal dysplasia protein) (EDA protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.006248 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.029540 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.007008 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.049126 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.019157 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
ADA2A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | -0.001106 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08913, Q2XN99, Q86TH8, Q9BZK1 | Gene names | ADRA2A, ADRA2R, ADRAR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR) (Alpha-2 adrenergic receptor subtype C10). | |||||
|
HASP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.014848 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0R0, Q99MU9, Q9JJJ4 | Gene names | Gsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (Germ cell-specific gene 2 protein). | |||||
|
HRBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.009597 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
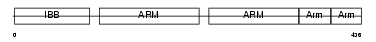
IMA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.119180 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
SHC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.006869 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.017375 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.056515 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
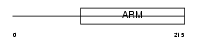
HEAT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.051002 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.118380 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.007458 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | -0.001391 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
AKAP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.009730 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JQC9, O60904, O95246, Q5JQD1, Q5JQD2, Q5JQD3 | Gene names | AKAP4, AKAP82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (hAKAP82) (Major sperm fibrous sheath protein) (HI). | |||||
|
ANR47_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.004579 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.046488 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.041406 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.010523 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.009906 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.001174 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NR4A3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.001970 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZB6, Q9QZB5 | Gene names | Nr4a3, Tec | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Orphan nuclear receptor TEC) (Translocated in extraskeletal chondrosarcoma). | |||||
|
NRX1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.003808 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
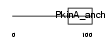
AKAP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.013556 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
ARRD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.007649 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
CADH4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.001067 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CLAP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.016102 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
COX10_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.009780 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
GCH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.011595 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.007915 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
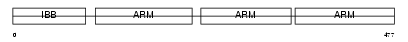
IMA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.105554 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
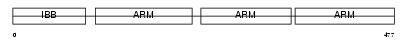
IMA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.105872 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.000768 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MAPK5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | -0.002652 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IW41, O60491, Q86X46, Q9BVX9, Q9UG86 | Gene names | MAPKAPK5, PRAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 5 (EC 2.7.11.1) (MAPK-activated protein kinase 5) (MAPKAP kinase 5) (p38-regulated/activated protein kinase). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.016075 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
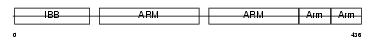
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.013109 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
CADH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.000849 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CCDC9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.007979 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.005976 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
IF38_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.009132 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
LYPD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.012758 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVP6 | Gene names | Lypd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ly6/PLAUR domain-containing protein 4 precursor. | |||||
|
NANO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.018894 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WY3, Q3UTS9, Q8BIJ9 | Gene names | Nanos1, Nos | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.004945 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
TRI56_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.003205 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
ZBED4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013447 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
ZNF12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | -0.002578 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
ARMC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.058872 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6L4 | Gene names | ARMC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMC7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.060259 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UJZ3, Q8R599 | Gene names | Armc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
GDS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.148364 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052603 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.995612 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.975107 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.974949 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.947803 (rank : 5) | θ value | 1.80368e-133 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.945767 (rank : 6) | θ value | 2.97994e-128 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.936130 (rank : 7) | θ value | 2.78268e-134 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.925315 (rank : 8) | θ value | 5.41814e-138 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
PKP2_HUMAN
|
||||||
| NC score | 0.917456 (rank : 9) | θ value | 3.96481e-64 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.910024 (rank : 10) | θ value | 5.17823e-64 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.909052 (rank : 11) | θ value | 8.83269e-64 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP1_MOUSE
|
||||||
| NC score | 0.905882 (rank : 12) | θ value | 1.84173e-61 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP1_HUMAN
|
||||||
| NC score | 0.904457 (rank : 13) | θ value | 1.41016e-61 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.405117 (rank : 14) | θ value | 5.81887e-07 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.323637 (rank : 15) | θ value | 3.41135e-07 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.323565 (rank : 16) | θ value | 3.41135e-07 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.289751 (rank : 17) | θ value | 1.09739e-05 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.251343 (rank : 18) | θ value | 0.00390308 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.237598 (rank : 19) | θ value | 0.000121331 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.231515 (rank : 20) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.148364 (rank : 21) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.131616 (rank : 22) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.130195 (rank : 23) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.130150 (rank : 24) | θ value | 0.163984 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.119180 (rank : 25) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.118380 (rank : 26) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.116274 (rank : 27) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.116203 (rank : 28) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.105872 (rank : 29) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.105554 (rank : 30) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.099336 (rank : 31) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.099217 (rank : 32) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
ARMC6_MOUSE
|
||||||
| NC score | 0.085220 (rank : 33) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
ARMC6_HUMAN
|
||||||
| NC score | 0.084574 (rank : 34) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
RTDR1_HUMAN
|
||||||
| NC score | 0.082229 (rank : 35) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
HEAT3_MOUSE
|
||||||
| NC score | 0.074819 (rank : 36) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.072186 (rank : 37) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.071548 (rank : 38) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMC7_MOUSE
|
||||||
| NC score | 0.060259 (rank : 39) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UJZ3, Q8R599 | Gene names | Armc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMC7_HUMAN
|
||||||
| NC score | 0.058872 (rank : 40) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6L4 | Gene names | ARMC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.056515 (rank : 41) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.052603 (rank : 42) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
HEAT3_HUMAN
|
||||||
| NC score | 0.051002 (rank : 43) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.049126 (rank : 44) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.046488 (rank : 45) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.041406 (rank : 46) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.037476 (rank : 47) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
PDLI4_HUMAN
|
||||||
| NC score | 0.036198 (rank : 48) | θ value | 0.0193708 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50479, Q96AT8, Q9BTW8, Q9Y292 | Gene names | PDLIM4, RIL | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
NANO1_HUMAN
|
||||||
| NC score | 0.034915 (rank : 49) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WY41 | Gene names | NANOS1, NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1 (NOS-1) (EC_Rep1a). | |||||
|
EDA_MOUSE
|
||||||
| NC score | 0.034255 (rank : 50) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54693, O35705, Q9QWJ8, Q9QZ01, Q9QZ02 | Gene names | Eda, Ed1, Ta | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (EDA protein homolog) (Tabby protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.029540 (rank : 51) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
XPC_MOUSE
|
||||||
| NC score | 0.027458 (rank : 52) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P51612, P54732, Q3TKI2, Q920M1, Q9DBW7 | Gene names | Xpc | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-C cells homolog (Xeroderma pigmentosum group C-complementing protein homolog) (p125). | |||||
|
EDA_HUMAN
|
||||||
| NC score | 0.026461 (rank : 53) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92838, O75910, Q5JUM7, Q9UP77, Q9Y6L0, Q9Y6L1, Q9Y6L2, Q9Y6L3, Q9Y6L4 | Gene names | EDA, ED1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectodysplasin-A (Ectodermal dysplasia protein) (EDA protein) [Contains: Ectodysplasin-A, membrane form; Ectodysplasin-A, secreted form]. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.025425 (rank : 54) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.021922 (rank : 55) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
CLAP2_HUMAN
|
||||||
| NC score | 0.020678 (rank : 56) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.019157 (rank : 57) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
NANO1_MOUSE
|
||||||
| NC score | 0.018894 (rank : 58) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WY3, Q3UTS9, Q8BIJ9 | Gene names | Nanos1, Nos | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1. | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.018368 (rank : 59) | θ value | 0.163984 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.017375 (rank : 60) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.016184 (rank : 61) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CLAP2_MOUSE
|
||||||
| NC score | 0.016102 (rank : 62) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.016075 (rank : 63) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
HASP_MOUSE
|
||||||
| NC score | 0.014848 (rank : 64) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z0R0, Q99MU9, Q9JJJ4 | Gene names | Gsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Haspin (EC 2.7.11.1) (Haploid germ cell-specific nuclear protein kinase) (Germ cell-specific gene 2 protein). | |||||
|
AKAP5_HUMAN
|
||||||
| NC score | 0.013556 (rank : 65) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
ZBED4_HUMAN
|
||||||
| NC score | 0.013447 (rank : 66) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.013109 (rank : 67) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.012791 (rank : 68) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
LYPD4_MOUSE
|
||||||
| NC score | 0.012758 (rank : 69) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVP6 | Gene names | Lypd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ly6/PLAUR domain-containing protein 4 precursor. | |||||
|
GCH1_HUMAN
|
||||||
| NC score | 0.011595 (rank : 70) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30793, Q9Y4I8 | Gene names | GCH1, DYT5, GCH | |||
|
Domain Architecture |

|
|||||
| Description | GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.010523 (rank : 71) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.009906 (rank : 72) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
COX10_MOUSE
|
||||||
| NC score | 0.009780 (rank : 73) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CFY5 | Gene names | Cox10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protoheme IX farnesyltransferase, mitochondrial precursor (EC 2.5.1.-) (Heme O synthase). | |||||
|
AKAP4_HUMAN
|
||||||
| NC score | 0.009730 (rank : 74) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JQC9, O60904, O95246, Q5JQD1, Q5JQD2, Q5JQD3 | Gene names | AKAP4, AKAP82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 4 precursor (Protein kinase A-anchoring protein 4) (PRKA4) (A-kinase anchor protein 82 kDa) (AKAP 82) (hAKAP82) (Major sperm fibrous sheath protein) (HI). | |||||
|
HRBL_MOUSE
|
||||||
| NC score | 0.009597 (rank : 75) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WC7, Q8BKS5, Q99J67 | Gene names | Hrbl, Rabr | |||
|
Domain Architecture |

|
|||||
| Description | HIV-1 Rev-binding protein-like protein (Rev/Rex activation domain- binding protein related) (RAB-R). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.009402 (rank : 76) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
IF38_HUMAN
|
||||||
| NC score | 0.009132 (rank : 77) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
CCDC9_MOUSE
|
||||||
| NC score | 0.007979 (rank : 78) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VC31 | Gene names | Ccdc9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 9. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.007915 (rank : 79) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
ARRD1_HUMAN
|
||||||
| NC score | 0.007649 (rank : 80) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N5I2 | Gene names | ARRDC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arrestin domain-containing protein 1. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.007458 (rank : 81) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.007008 (rank : 82) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
SHC1_MOUSE
|
||||||
| NC score | 0.006869 (rank : 83) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98083, Q3U2Q7, Q8BFY3, Q8K4C6, Q8K4C7 | Gene names | Shc1, Shc, ShcA | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 1 (SH2 domain protein C1) (Src homology 2 domain-containing-transforming protein C1). | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.006248 (rank : 84) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.005976 (rank : 85) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.004945 (rank : 86) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ANR47_MOUSE
|
||||||
| NC score | 0.004579 (rank : 87) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1P7 | Gene names | Ankrd47, D17Ertd288e, Ng28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 47. | |||||
|
NRX1A_HUMAN
|
||||||
| NC score | 0.003808 (rank : 88) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULB1, O60323, Q53TJ9, Q53TQ1, Q9C079, Q9C080, Q9C081, Q9H3M2, Q9UDM6 | Gene names | NRXN1, KIAA0578 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-1-alpha precursor (Neurexin I-alpha). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.003205 (rank : 89) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
NR4A3_MOUSE
|
||||||
| NC score | 0.001970 (rank : 90) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZB6, Q9QZB5 | Gene names | Nr4a3, Tec | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Orphan nuclear receptor TEC) (Translocated in extraskeletal chondrosarcoma). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.001174 (rank : 91) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
CADH4_HUMAN
|
||||||
| NC score | 0.001067 (rank : 92) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH4_MOUSE
|
||||||
| NC score | 0.000849 (rank : 93) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.000768 (rank : 94) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KLF13_MOUSE
|
||||||
| NC score | 0.000445 (rank : 95) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JJZ6, Q9ESX3, Q9JHF8 | Gene names | Klf13, Bteb3, Fklf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 13 (Transcription factor BTEB3) (Basic transcription element-binding protein 3) (BTE-binding protein 3) (RANTES factor of late activated T-lymphocytes 1) (RFLAT-1) (Erythroid transcription factor FKLF-2). | |||||
|
ADA2A_HUMAN
|
||||||
| NC score | -0.001106 (rank : 96) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08913, Q2XN99, Q86TH8, Q9BZK1 | Gene names | ADRA2A, ADRA2R, ADRAR | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-2A adrenergic receptor (Alpha-2A adrenoceptor) (Alpha-2A adrenoreceptor) (Alpha-2AAR) (Alpha-2 adrenergic receptor subtype C10). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | -0.001391 (rank : 97) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ZNF12_HUMAN
|
||||||
| NC score | -0.002578 (rank : 98) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
MAPK5_HUMAN
|
||||||
| NC score | -0.002652 (rank : 99) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IW41, O60491, Q86X46, Q9BVX9, Q9UG86 | Gene names | MAPKAPK5, PRAK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAP kinase-activated protein kinase 5 (EC 2.7.11.1) (MAPK-activated protein kinase 5) (MAPKAP kinase 5) (p38-regulated/activated protein kinase). | |||||