Please be patient as the page loads
|
ARVC_MOUSE
|
||||||
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARVC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.995612 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.977265 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.977316 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 1.68428e-139 (rank : 5) | NC score | 0.927198 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 3.28708e-135 (rank : 6) | NC score | 0.938610 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 5.80223e-132 (rank : 7) | NC score | 0.949704 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 3.64273e-126 (rank : 8) | NC score | 0.947461 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP2_HUMAN
|
||||||
| θ value | 1.45591e-66 (rank : 9) | NC score | 0.920850 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 3.35642e-63 (rank : 10) | NC score | 0.912157 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| θ value | 5.7252e-63 (rank : 11) | NC score | 0.911166 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP1_HUMAN
|
||||||
| θ value | 7.47731e-63 (rank : 12) | NC score | 0.907528 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP1_MOUSE
|
||||||
| θ value | 7.47731e-63 (rank : 13) | NC score | 0.909203 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 14) | NC score | 0.406518 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 15) | NC score | 0.325688 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 16) | NC score | 0.325758 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 17) | NC score | 0.292820 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
APC_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.231155 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
APC_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 19) | NC score | 0.237044 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
PLAK_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 20) | NC score | 0.254301 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 21) | NC score | 0.114685 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
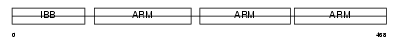
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.114757 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
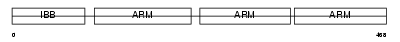
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
PDLI4_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.035464 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50479, Q96AT8, Q9BTW8, Q9Y292 | Gene names | PDLIM4, RIL | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.030373 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.075899 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.129360 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
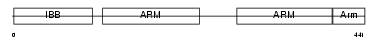
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
ARMC6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.085210 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.129298 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.075201 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMC6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.084757 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.037508 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.098213 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.018072 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
IMA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.098351 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
RTDR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.082317 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.130813 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
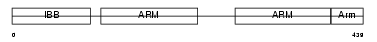
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
HEAT3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.074870 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.003893 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
LSR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.019262 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.022075 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
AATK_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.000930 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.011030 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.051366 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.010897 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.118543 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.117742 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.058770 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.004102 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
HEAT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.051074 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |
|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.012956 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.045779 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.043856 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | -0.000058 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.010109 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.001913 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.002763 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.006235 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
CSTF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.003237 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |
|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
ERGI2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.006707 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CR89, Q3UX13, Q9CWM6, Q9CYA2, Q9D4R1, Q9D8Z9 | Gene names | Ergic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endoplasmic reticulum-Golgi intermediate compartment protein 2. | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.104067 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.104330 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
LYPD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.012782 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVP6 | Gene names | Lypd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ly6/PLAUR domain-containing protein 4 precursor. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.029384 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.005049 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRPV5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.001983 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
ZBED4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.013476 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
ZNF12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | -0.002878 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||
|
ARMC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.059419 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6L4 | Gene names | ARMC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMC7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.060888 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UJZ3, Q8R599 | Gene names | Armc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
GDS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.149017 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.051448 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.995612 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.977316 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.977265 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.949704 (rank : 5) | θ value | 5.80223e-132 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.947461 (rank : 6) | θ value | 3.64273e-126 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.938610 (rank : 7) | θ value | 3.28708e-135 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.927198 (rank : 8) | θ value | 1.68428e-139 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
PKP2_HUMAN
|
||||||
| NC score | 0.920850 (rank : 9) | θ value | 1.45591e-66 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.912157 (rank : 10) | θ value | 3.35642e-63 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.911166 (rank : 11) | θ value | 5.7252e-63 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP1_MOUSE
|
||||||
| NC score | 0.909203 (rank : 12) | θ value | 7.47731e-63 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP1_HUMAN
|
||||||
| NC score | 0.907528 (rank : 13) | θ value | 7.47731e-63 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.406518 (rank : 14) | θ value | 7.59969e-07 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.325758 (rank : 15) | θ value | 7.59969e-07 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.325688 (rank : 16) | θ value | 7.59969e-07 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.292820 (rank : 17) | θ value | 2.21117e-06 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.254301 (rank : 18) | θ value | 0.00102713 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.237044 (rank : 19) | θ value | 0.000121331 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.231155 (rank : 20) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.149017 (rank : 21) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.130813 (rank : 22) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.129360 (rank : 23) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.129298 (rank : 24) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.118543 (rank : 25) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.117742 (rank : 26) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.114757 (rank : 27) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.114685 (rank : 28) | θ value | 0.0431538 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.104330 (rank : 29) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.104067 (rank : 30) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.098351 (rank : 31) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.098213 (rank : 32) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
ARMC6_MOUSE
|
||||||
| NC score | 0.085210 (rank : 33) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BNU0, Q8C4P5, Q8C7S1, Q8K2S7, Q99JQ2, Q9CWE4 | Gene names | Armc6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
ARMC6_HUMAN
|
||||||
| NC score | 0.084757 (rank : 34) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NXE6, O94999, Q9BTH5 | Gene names | ARMC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 6. | |||||
|
RTDR1_HUMAN
|
||||||
| NC score | 0.082317 (rank : 35) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.075899 (rank : 36) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.075201 (rank : 37) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
HEAT3_MOUSE
|
||||||
| NC score | 0.074870 (rank : 38) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BQM4, Q8C731 | Gene names | Heatr3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
ARMC7_MOUSE
|
||||||
| NC score | 0.060888 (rank : 39) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q3UJZ3, Q8R599 | Gene names | Armc7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMC7_HUMAN
|
||||||
| NC score | 0.059419 (rank : 40) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H6L4 | Gene names | ARMC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 7. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.058770 (rank : 41) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.051448 (rank : 42) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.051366 (rank : 43) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
HEAT3_HUMAN
|
||||||
| NC score | 0.051074 (rank : 44) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z4Q2, Q8N525, Q8WV56, Q96CC9, Q9NWN7 | Gene names | HEATR3 | |||
|
Domain Architecture |

|
|||||
| Description | HEAT repeat-containing protein 3. | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.045779 (rank : 45) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.043856 (rank : 46) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.037508 (rank : 47) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
PDLI4_HUMAN
|
||||||
| NC score | 0.035464 (rank : 48) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P50479, Q96AT8, Q9BTW8, Q9Y292 | Gene names | PDLIM4, RIL | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.030373 (rank : 49) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.029384 (rank : 50) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.022075 (rank : 51) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
LSR_HUMAN
|
||||||
| NC score | 0.019262 (rank : 52) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X29, O00112, O00426, Q6ZT80, Q9UQL3 | Gene names | LSR, LISCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipolysis-stimulated lipoprotein receptor. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.018072 (rank : 53) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZBED4_HUMAN
|
||||||
| NC score | 0.013476 (rank : 54) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75132, Q9UGG8 | Gene names | ZBED4, KIAA0637 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger BED domain-containing protein 4. | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.012956 (rank : 55) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
LYPD4_MOUSE
|
||||||
| NC score | 0.012782 (rank : 56) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVP6 | Gene names | Lypd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ly6/PLAUR domain-containing protein 4 precursor. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.011030 (rank : 57) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.010897 (rank : 58) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.010109 (rank : 59) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
ERGI2_MOUSE
|
||||||
| NC score | 0.006707 (rank : 60) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CR89, Q3UX13, Q9CWM6, Q9CYA2, Q9D4R1, Q9D8Z9 | Gene names | Ergic2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endoplasmic reticulum-Golgi intermediate compartment protein 2. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.006235 (rank : 61) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.005049 (rank : 62) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.004102 (rank : 63) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.003893 (rank : 64) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CSTF2_HUMAN
|
||||||
| NC score | 0.003237 (rank : 65) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P33240, Q5H951, Q6LA74, Q8N502 | Gene names | CSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage stimulation factor 64 kDa subunit (CSTF 64 kDa subunit) (CF-1 64 kDa subunit) (CstF-64). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.002763 (rank : 66) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TRPV5_HUMAN
|
||||||
| NC score | 0.001983 (rank : 67) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQA5, Q8NDW5, Q8NDX7, Q8NDX8, Q96PM6 | Gene names | TRPV5, ECAC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 5 (TrpV5) (osm-9-like TRP channel 3) (OTRPC3) (Epithelial calcium channel 1) (ECaC) (ECaC1) (Calcium transport protein 2) (CaT2). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.001913 (rank : 68) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.000930 (rank : 69) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | -0.000058 (rank : 70) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ZNF12_HUMAN
|
||||||
| NC score | -0.002878 (rank : 71) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17014, Q9ULZ6 | Gene names | ZNF12, GIOT3, KOX3, ZNF325 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 12 (Zinc finger protein KOX3) (Zinc finger protein 325) (Gonadotropin-inducible transcription repressor 3) (GIOT-3). | |||||