Please be patient as the page loads
|
IMA4_MOUSE
|
||||||
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
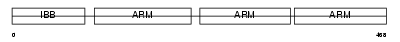
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IMA3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996484 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
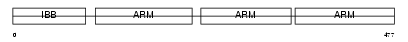
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996410 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
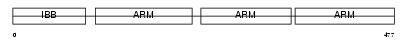
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999827 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
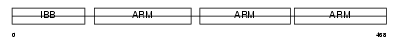
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 1.47209e-143 (rank : 5) | NC score | 0.977093 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
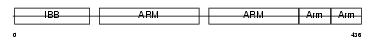
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 3.62588e-142 (rank : 6) | NC score | 0.975346 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
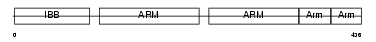
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 5.43074e-130 (rank : 7) | NC score | 0.970373 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 1.33764e-128 (rank : 8) | NC score | 0.972265 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
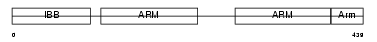
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 1.74701e-128 (rank : 9) | NC score | 0.973148 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
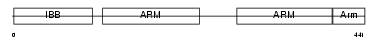
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 1.69212e-123 (rank : 10) | NC score | 0.971320 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA1_HUMAN
|
||||||
| θ value | 2.20997e-123 (rank : 11) | NC score | 0.971792 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 12) | NC score | 0.635506 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 13) | NC score | 0.634731 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.161978 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 15) | NC score | 0.162253 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 16) | NC score | 0.114031 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.157389 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.157433 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
TNPO2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.106366 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.107614 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 21) | NC score | 0.116274 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
SARM1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 22) | NC score | 0.112392 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.114757 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
SARM1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 24) | NC score | 0.112067 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
GCN1L_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.084320 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.012423 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TNPO1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.120712 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
TNPO1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.115784 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
GDS1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.265697 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.054952 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.119142 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
IMB3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.050359 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00410, O15257 | Gene names | RANBP5, KPNB3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
IMB3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.050466 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BKC5, Q3TEG2, Q7TQC6, Q9EQ30 | Gene names | Ranbp5, Kpnb3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.111381 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
SIL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.049541 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H173, Q8N2L3 | Gene names | SIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor (BiP-associated protein) (BAP). | |||||
|
ZC3H6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.021604 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.123697 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.124678 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
IMB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.100625 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.013746 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
PHTF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.024290 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ09, Q9CS51, Q9CSB9, Q9QZ14 | Gene names | Phtf1, Phtf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative homeodomain transcription factor 1. | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.054813 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.045071 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.046937 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.233555 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.007985 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.031433 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.028948 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.012257 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
REST_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.007043 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RIF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.014865 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.003075 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.004922 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
DHX16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.004961 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
PKP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.061203 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
MMS19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.012609 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D071, Q925N8, Q9CWX3 | Gene names | Mms19l, Mms19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (MET18 homolog). | |||||
|
NF2L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.006812 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
PTN7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.005367 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
TTMP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.009200 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C5C9, Q91YJ8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TPA-induced transmembrane protein homolog. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.012296 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
IMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.079305 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
PKP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.056968 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.054851 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.052278 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PLAK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.101336 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.133167 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
RL35_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.085039 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42766, Q4VBY5, Q5JTN5, Q6IBC7, Q96QJ7, Q9BYF4 | Gene names | RPL35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L35. | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.999827 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.996484 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.996410 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.977093 (rank : 5) | θ value | 1.47209e-143 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.975346 (rank : 6) | θ value | 3.62588e-142 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.973148 (rank : 7) | θ value | 1.74701e-128 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.972265 (rank : 8) | θ value | 1.33764e-128 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.971792 (rank : 9) | θ value | 2.20997e-123 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.971320 (rank : 10) | θ value | 1.69212e-123 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.970373 (rank : 11) | θ value | 5.43074e-130 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.635506 (rank : 12) | θ value | 1.29331e-14 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.634731 (rank : 13) | θ value | 2.88119e-14 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.265697 (rank : 14) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.233555 (rank : 15) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.162253 (rank : 16) | θ value | 0.00509761 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.161978 (rank : 17) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.157433 (rank : 18) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.157389 (rank : 19) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.133167 (rank : 20) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.124678 (rank : 21) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.123697 (rank : 22) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
TNPO1_HUMAN
|
||||||
| NC score | 0.120712 (rank : 23) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.119142 (rank : 24) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.116274 (rank : 25) | θ value | 0.0330416 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
TNPO1_MOUSE
|
||||||
| NC score | 0.115784 (rank : 26) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.114757 (rank : 27) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.114031 (rank : 28) | θ value | 0.00665767 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
SARM1_MOUSE
|
||||||
| NC score | 0.112392 (rank : 29) | θ value | 0.0330416 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.112067 (rank : 30) | θ value | 0.0431538 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.111381 (rank : 31) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
TNPO2_MOUSE
|
||||||
| NC score | 0.107614 (rank : 32) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_HUMAN
|
||||||
| NC score | 0.106366 (rank : 33) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.101336 (rank : 34) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
IMB1_MOUSE
|
||||||
| NC score | 0.100625 (rank : 35) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
RL35_HUMAN
|
||||||
| NC score | 0.085039 (rank : 36) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42766, Q4VBY5, Q5JTN5, Q6IBC7, Q96QJ7, Q9BYF4 | Gene names | RPL35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L35. | |||||
|
GCN1L_HUMAN
|
||||||
| NC score | 0.084320 (rank : 37) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
IMB1_HUMAN
|
||||||
| NC score | 0.079305 (rank : 38) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
PKP1_HUMAN
|
||||||
| NC score | 0.061203 (rank : 39) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP1_MOUSE
|
||||||
| NC score | 0.056968 (rank : 40) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.054952 (rank : 41) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.054851 (rank : 42) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.054813 (rank : 43) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.052278 (rank : 44) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
IMB3_MOUSE
|
||||||
| NC score | 0.050466 (rank : 45) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BKC5, Q3TEG2, Q7TQC6, Q9EQ30 | Gene names | Ranbp5, Kpnb3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
IMB3_HUMAN
|
||||||
| NC score | 0.050359 (rank : 46) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00410, O15257 | Gene names | RANBP5, KPNB3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
SIL1_HUMAN
|
||||||
| NC score | 0.049541 (rank : 47) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H173, Q8N2L3 | Gene names | SIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleotide exchange factor SIL1 precursor (BiP-associated protein) (BAP). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.046937 (rank : 48) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.045071 (rank : 49) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.031433 (rank : 50) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.028948 (rank : 51) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
PHTF1_MOUSE
|
||||||
| NC score | 0.024290 (rank : 52) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZ09, Q9CS51, Q9CSB9, Q9QZ14 | Gene names | Phtf1, Phtf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative homeodomain transcription factor 1. | |||||
|
ZC3H6_MOUSE
|
||||||
| NC score | 0.021604 (rank : 53) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYK8, Q80UK3, Q8C1H1, Q9D604 | Gene names | Zc3h6, Zc3hdc6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
RIF1_MOUSE
|
||||||
| NC score | 0.014865 (rank : 54) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PR54, Q6T8D9, Q7TPD8, Q8BTU2, Q8C408, Q9CS77 | Gene names | Rif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog) (mRif1). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.013746 (rank : 55) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MMS19_MOUSE
|
||||||
| NC score | 0.012609 (rank : 56) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D071, Q925N8, Q9CWX3 | Gene names | Mms19l, Mms19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (MET18 homolog). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.012423 (rank : 57) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.012296 (rank : 58) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.012257 (rank : 59) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
TTMP_MOUSE
|
||||||
| NC score | 0.009200 (rank : 60) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C5C9, Q91YJ8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TPA-induced transmembrane protein homolog. | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.007985 (rank : 61) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.007043 (rank : 62) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
NF2L2_MOUSE
|
||||||
| NC score | 0.006812 (rank : 63) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60795 | Gene names | Nfe2l2, Nrf-2, Nrf2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2). | |||||
|
PTN7_HUMAN
|
||||||
| NC score | 0.005367 (rank : 64) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35236, Q9BV05 | Gene names | PTPN7 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 7 (EC 3.1.3.48) (Protein-tyrosine phosphatase LC-PTP) (Hematopoietic protein-tyrosine phosphatase) (HEPTP). | |||||
|
DHX16_HUMAN
|
||||||
| NC score | 0.004961 (rank : 65) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60231, O60322, Q969X7, Q96QC1 | Gene names | DHX16, DBP2, DDX16, KIAA0577 | |||
|
Domain Architecture |

|
|||||
| Description | Putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 (EC 3.6.1.-) (DEAH-box protein 16) (ATP-dependent RNA helicase #3). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.004922 (rank : 66) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.003075 (rank : 67) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||