Please be patient as the page loads
|
IMA5_HUMAN
|
||||||
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IMA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991482 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991340 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA7_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992762 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |
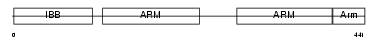
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA7_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.991276 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |
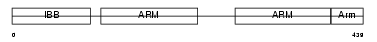
|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 1.92707e-135 (rank : 6) | NC score | 0.972133 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
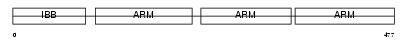
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 3.6343e-134 (rank : 7) | NC score | 0.972039 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
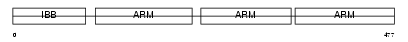
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 6.41512e-131 (rank : 8) | NC score | 0.970810 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
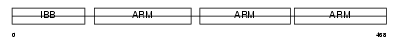
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 5.43074e-130 (rank : 9) | NC score | 0.970373 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
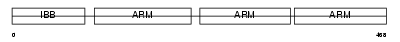
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | 1.13238e-127 (rank : 10) | NC score | 0.974478 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
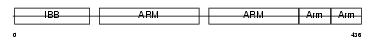
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA2_HUMAN
|
||||||
| θ value | 3.64273e-126 (rank : 11) | NC score | 0.972242 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
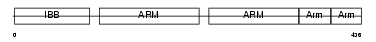
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 12) | NC score | 0.640789 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 13) | NC score | 0.637872 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
ARMC4_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 14) | NC score | 0.286632 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
GDS1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.306536 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 16) | NC score | 0.085504 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 17) | NC score | 0.084915 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
IMB1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 18) | NC score | 0.106684 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 19) | NC score | 0.175562 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 20) | NC score | 0.175603 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 21) | NC score | 0.145200 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
IMB1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 22) | NC score | 0.127826 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
PKP4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 23) | NC score | 0.175243 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
PKP4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 24) | NC score | 0.177495 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 25) | NC score | 0.145260 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 26) | NC score | 0.078134 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
TNPO1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 27) | NC score | 0.099123 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
TNPO1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.095373 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
LRRK2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 29) | NC score | 0.006957 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5S006, Q8BWG7, Q8BZJ6, Q8CI84, Q8K062 | Gene names | Lrrk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PLAK_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 30) | NC score | 0.175290 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.069644 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARVC_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.130195 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 33) | NC score | 0.129298 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
CTND1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.122122 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
CTND1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.130756 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.059043 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
ZFY16_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.036438 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
PLAK_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.138087 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
DBNL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 39) | NC score | 0.017423 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.011461 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
AGGF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.017551 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
RELN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.046333 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
2AAA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.022641 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30153, Q13773 | Gene names | PPP2R1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein). | |||||
|
RELN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.034327 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |
|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.000022 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.048624 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
PERQ1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.017142 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR1 | Gene names | Perq1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
RL35_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.116205 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42766, Q4VBY5, Q5JTN5, Q6IBC7, Q96QJ7, Q9BYF4 | Gene names | RPL35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L35. | |||||
|
K1949_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.016268 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
PKP3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.069655 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.008923 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
2AAA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.020783 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q76MZ3 | Gene names | Ppp2r1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform). | |||||
|
TBCC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.015931 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15814 | Gene names | TBCC | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin-specific chaperone C (Tubulin-folding cofactor C) (CFC). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.016328 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.013205 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
DBNL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.012940 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJU6, P84070, Q6IAI8, Q96F30, Q96K74, Q9HBN8, Q9NR72 | Gene names | DBNL, CMAP, SH3P7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.012620 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | -0.001012 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
APC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.037999 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
APC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.039268 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.000848 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CLAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.015190 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.005538 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PKP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.066877 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
CF081_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.031336 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
K1219_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.006169 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.004854 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
PRGC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.005994 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
CF081_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.060509 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
GCN1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052751 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
PKP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.068448 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.065666 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
PKP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.061051 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
SARM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.061918 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SARM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.062747 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
TNPO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.081527 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.082964 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.091522 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
IMA7_HUMAN
|
||||||
| NC score | 0.992762 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O60684 | Gene names | KPNA6, IPOA7 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6). | |||||
|
IMA1_HUMAN
|
||||||
| NC score | 0.991482 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P52294, Q9BQ56 | Gene names | KPNA1, RCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (NPI-1). | |||||
|
IMA1_MOUSE
|
||||||
| NC score | 0.991340 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q60960, Q3TF32 | Gene names | Kpna1, Rch2 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-1 subunit (Karyopherin alpha-1 subunit) (SRP1-beta) (RAG cohort protein 2) (Nucleoprotein interactor 1) (Importin alpha S1). | |||||
|
IMA7_MOUSE
|
||||||
| NC score | 0.991276 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O35345, Q9CVP9 | Gene names | Kpna6, Kpna5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-7 subunit (Karyopherin alpha-6 subunit) (Importin alpha S2). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.974478 (rank : 6) | θ value | 1.13238e-127 (rank : 10) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.972242 (rank : 7) | θ value | 3.64273e-126 (rank : 11) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.972133 (rank : 8) | θ value | 1.92707e-135 (rank : 6) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.972039 (rank : 9) | θ value | 3.6343e-134 (rank : 7) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.970810 (rank : 10) | θ value | 6.41512e-131 (rank : 8) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.970373 (rank : 11) | θ value | 5.43074e-130 (rank : 9) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.640789 (rank : 12) | θ value | 8.95645e-16 (rank : 12) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.637872 (rank : 13) | θ value | 4.44505e-15 (rank : 13) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
GDS1_HUMAN
|
||||||
| NC score | 0.306536 (rank : 14) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52306, Q9NYM2, Q9NZA8 | Gene names | RAP1GDS1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-GDP dissociation stimulator 1 (SMG P21 stimulatory GDP/GTP exchange protein) (SMG GDS protein) (Exchange factor smgGDS). | |||||
|
ARMC4_HUMAN
|
||||||
| NC score | 0.286632 (rank : 15) | θ value | 1.43324e-05 (rank : 14) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5T2S8, Q9H0C0 | Gene names | ARMC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing protein 4. | |||||
|
PKP4_MOUSE
|
||||||
| NC score | 0.177495 (rank : 16) | θ value | 0.0148317 (rank : 24) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q68FH0, Q640N0, Q68G56, Q8BK47, Q8BVH1, Q9CRE3 | Gene names | Pkp4, Armrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plakophilin-4 (Armadillo-related protein). | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.175603 (rank : 17) | θ value | 0.0113563 (rank : 20) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.175562 (rank : 18) | θ value | 0.0113563 (rank : 19) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.175290 (rank : 19) | θ value | 0.0736092 (rank : 30) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PKP4_HUMAN
|
||||||
| NC score | 0.175243 (rank : 20) | θ value | 0.0148317 (rank : 23) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99569 | Gene names | PKP4 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-4 (p0071). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.145260 (rank : 21) | θ value | 0.0193708 (rank : 25) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.145200 (rank : 22) | θ value | 0.0148317 (rank : 21) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.138087 (rank : 23) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
CTND1_MOUSE
|
||||||
| NC score | 0.130756 (rank : 24) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P30999 | Gene names | Ctnnd1, Catns | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
ARVC_HUMAN
|
||||||
| NC score | 0.130195 (rank : 25) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O00192 | Gene names | ARVCF | |||
|
Domain Architecture |

|
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome. | |||||
|
ARVC_MOUSE
|
||||||
| NC score | 0.129298 (rank : 26) | θ value | 0.21417 (rank : 33) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98203, Q6PGJ6, Q8BQ36, Q8BRF2, Q8C3U7, Q924L2, Q924L3, Q924L4, Q924L5 | Gene names | Arvcf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat protein deleted in velo-cardio-facial syndrome homolog. | |||||
|
IMB1_MOUSE
|
||||||
| NC score | 0.127826 (rank : 27) | θ value | 0.0148317 (rank : 22) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
CTND1_HUMAN
|
||||||
| NC score | 0.122122 (rank : 28) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60716, O15088, O60713, O60714, O60715, O60935, Q6RBX8, Q9UP71, Q9UP72, Q9UP73 | Gene names | CTNND1, KIAA0384 | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-1 (p120 catenin) (p120(ctn)) (Cadherin-associated Src substrate) (CAS) (p120(cas)). | |||||
|
RL35_HUMAN
|
||||||
| NC score | 0.116205 (rank : 29) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P42766, Q4VBY5, Q5JTN5, Q6IBC7, Q96QJ7, Q9BYF4 | Gene names | RPL35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 60S ribosomal protein L35. | |||||
|
IMB1_HUMAN
|
||||||
| NC score | 0.106684 (rank : 30) | θ value | 0.00869519 (rank : 18) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
TNPO1_HUMAN
|
||||||
| NC score | 0.099123 (rank : 31) | θ value | 0.0563607 (rank : 27) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
TNPO1_MOUSE
|
||||||
| NC score | 0.095373 (rank : 32) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.091522 (rank : 33) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.085504 (rank : 34) | θ value | 0.00228821 (rank : 16) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.084915 (rank : 35) | θ value | 0.00298849 (rank : 17) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
TNPO2_MOUSE
|
||||||
| NC score | 0.082964 (rank : 36) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_HUMAN
|
||||||
| NC score | 0.081527 (rank : 37) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.078134 (rank : 38) | θ value | 0.0563607 (rank : 26) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
PKP3_HUMAN
|
||||||
| NC score | 0.069655 (rank : 39) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y446 | Gene names | PKP3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.069644 (rank : 40) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
PKP1_HUMAN
|
||||||
| NC score | 0.068448 (rank : 41) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13835, O00645, Q15152 | Gene names | PKP1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1 (Band-6 protein) (B6P). | |||||
|
PKP3_MOUSE
|
||||||
| NC score | 0.066877 (rank : 42) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QY23 | Gene names | Pkp3 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-3. | |||||
|
PKP1_MOUSE
|
||||||
| NC score | 0.065666 (rank : 43) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97350 | Gene names | Pkp1 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-1. | |||||
|
SARM1_MOUSE
|
||||||
| NC score | 0.062747 (rank : 44) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PDS3, Q5SYG5, Q5SYG6, Q6A054, Q6SZW0, Q8BRI9 | Gene names | Sarm1, Kiaa0524 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Tir-1 homolog). | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.061918 (rank : 45) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
PKP2_HUMAN
|
||||||
| NC score | 0.061051 (rank : 46) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99959, Q99960 | Gene names | PKP2 | |||
|
Domain Architecture |

|
|||||
| Description | Plakophilin-2. | |||||
|
CF081_HUMAN
|
||||||
| NC score | 0.060509 (rank : 47) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.059043 (rank : 48) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
GCN1L_HUMAN
|
||||||
| NC score | 0.052751 (rank : 49) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.048624 (rank : 50) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
RELN_MOUSE
|
||||||
| NC score | 0.046333 (rank : 51) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60841, Q9CUA6 | Gene names | Reln, Rl | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-) (Reeler protein). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.039268 (rank : 52) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
APC_HUMAN
|
||||||
| NC score | 0.037999 (rank : 53) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 603 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P25054, Q15162, Q15163, Q93042 | Gene names | APC, DP2.5 | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC). | |||||
|
ZFY16_HUMAN
|
||||||
| NC score | 0.036438 (rank : 54) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z3T8, O15023, Q7LAU7, Q86T69, Q8N5L3, Q8NEK3 | Gene names | ZFYVE16, KIAA0305 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 16 (Endofin) (Endosome- associated FYVE domain protein). | |||||
|
RELN_HUMAN
|
||||||
| NC score | 0.034327 (rank : 55) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78509, Q86UJ0, Q86UJ8, Q8NDV0, Q9UDQ2 | Gene names | RELN | |||
|
Domain Architecture |

|
|||||
| Description | Reelin precursor (EC 3.4.21.-). | |||||
|
CF081_MOUSE
|
||||||
| NC score | 0.031336 (rank : 56) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
2AAA_HUMAN
|
||||||
| NC score | 0.022641 (rank : 57) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P30153, Q13773 | Gene names | PPP2R1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein). | |||||
|
2AAA_MOUSE
|
||||||
| NC score | 0.020783 (rank : 58) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q76MZ3 | Gene names | Ppp2r1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform). | |||||
|
AGGF1_HUMAN
|
||||||
| NC score | 0.017551 (rank : 59) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N302, O00581, Q53YS3, Q9BU84, Q9NW66 | Gene names | AGGF1, VG5Q | |||
|
Domain Architecture |

|
|||||
| Description | Angiogenic factor with G patch and FHA domains 1 (Angiogenic factor VG5Q) (Vasculogenesis gene on 5q protein) (hVG5Q). | |||||
|
DBNL_MOUSE
|
||||||
| NC score | 0.017423 (rank : 60) | θ value | 0.62314 (rank : 39) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q62418, Q80WP1, Q8BH56 | Gene names | Dbnl, Abp1, Sh3p7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Actin-binding protein 1). | |||||
|
PERQ1_MOUSE
|
||||||
| NC score | 0.017142 (rank : 61) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MR1 | Gene names | Perq1 | |||
|
Domain Architecture |

|
|||||
| Description | PERQ amino acid rich with GYF domain protein 1. | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.016328 (rank : 62) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
K1949_HUMAN
|
||||||
| NC score | 0.016268 (rank : 63) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NYC8, Q68CK8, Q6ZUJ6, Q8NDQ4, Q8TF52, Q9BRL9 | Gene names | KIAA1949 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1949. | |||||
|
TBCC_HUMAN
|
||||||
| NC score | 0.015931 (rank : 64) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15814 | Gene names | TBCC | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin-specific chaperone C (Tubulin-folding cofactor C) (CFC). | |||||
|
CLAP1_HUMAN
|
||||||
| NC score | 0.015190 (rank : 65) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.013205 (rank : 66) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
DBNL_HUMAN
|
||||||
| NC score | 0.012940 (rank : 67) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJU6, P84070, Q6IAI8, Q96F30, Q96K74, Q9HBN8, Q9NR72 | Gene names | DBNL, CMAP, SH3P7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Drebrin-like protein (SH3 domain-containing protein 7) (Drebrin F) (Cervical SH3P7) (HPK1-interacting protein of 55 kDa) (HIP-55) (Cervical mucin-associated protein). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.012620 (rank : 68) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.011461 (rank : 69) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.008923 (rank : 70) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
LRRK2_MOUSE
|
||||||
| NC score | 0.006957 (rank : 71) | θ value | 0.0736092 (rank : 29) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5S006, Q8BWG7, Q8BZJ6, Q8CI84, Q8K062 | Gene names | Lrrk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
K1219_HUMAN
|
||||||
| NC score | 0.006169 (rank : 72) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
PRGC1_HUMAN
|
||||||
| NC score | 0.005994 (rank : 73) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBK2, Q9UN32 | Gene names | PPARGC1A, LEM6, PGC1, PGC1A, PPARGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPAR gamma coactivator 1-alpha) (PPARGC-1-alpha) (PGC-1-alpha) (Ligand effect modulator 6). | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.005538 (rank : 74) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.004854 (rank : 75) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.000848 (rank : 76) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.000022 (rank : 77) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
WNK1_HUMAN
|
||||||
| NC score | -0.001012 (rank : 78) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 68 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||