Please be patient as the page loads
|
TBCC_HUMAN
|
||||||
| SwissProt Accessions | Q15814 | Gene names | TBCC | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin-specific chaperone C (Tubulin-folding cofactor C) (CFC). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TBCC_HUMAN
|
||||||
| θ value | 6.59259e-152 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15814 | Gene names | TBCC | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin-specific chaperone C (Tubulin-folding cofactor C) (CFC). | |||||
|
XRP2_HUMAN
|
||||||
| θ value | 2.20094e-22 (rank : 2) | NC score | 0.726885 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75695, Q86XJ7, Q9NU67 | Gene names | RP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein XRP2. | |||||
|
XRP2_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 3) | NC score | 0.724493 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPK2, Q8BLN8, Q8BVQ8, Q8BZP9 | Gene names | Rp2, Rp2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein XRP2. | |||||
|
K1H2_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.052872 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |
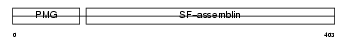
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
KIFC3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 5) | NC score | 0.060870 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
KIFC3_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.061635 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 571 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35231, Q91YQ2 | Gene names | Kifc3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
K1H7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.044928 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76014 | Gene names | KRTHA7, HHA7, HKA7 | |||
|
Domain Architecture |
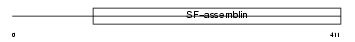
|
|||||
| Description | Keratin, type I cuticular Ha7 (Hair keratin, type I Ha7). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.058925 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.044722 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.044035 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
K1H1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.041750 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61765 | Gene names | Krtha1, Hka1, Krt1-1 | |||
|
Domain Architecture |
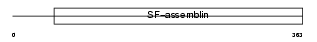
|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1) (HKA-1). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.037905 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CING_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.048759 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.039099 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.040297 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
XRCC4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.044484 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
MY18A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.038532 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.041612 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.038628 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
HGS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.036584 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.036673 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
IMA5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.015931 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
A4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.033012 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.032908 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.033620 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.050679 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.020262 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.020090 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
CING_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.046375 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.037538 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.038817 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RPB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.044772 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15514, Q52LT4 | Gene names | POLR2D | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II 16 kDa polypeptide (EC 2.7.7.6) (RPB4). | |||||
|
RPB4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.044772 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7M8, Q6ZWW0, Q78KE6 | Gene names | Polr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase II 16 kDa polypeptide (EC 2.7.7.6) (RPB4). | |||||
|
BRE1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.039891 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
BRE1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.038472 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.032473 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
STIM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.034367 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.034349 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TBCC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.59259e-152 (rank : 1) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15814 | Gene names | TBCC | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin-specific chaperone C (Tubulin-folding cofactor C) (CFC). | |||||
|
XRP2_HUMAN
|
||||||
| NC score | 0.726885 (rank : 2) | θ value | 2.20094e-22 (rank : 2) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75695, Q86XJ7, Q9NU67 | Gene names | RP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein XRP2. | |||||
|
XRP2_MOUSE
|
||||||
| NC score | 0.724493 (rank : 3) | θ value | 1.09232e-21 (rank : 3) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPK2, Q8BLN8, Q8BVQ8, Q8BZP9 | Gene names | Rp2, Rp2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein XRP2. | |||||
|
KIFC3_MOUSE
|
||||||
| NC score | 0.061635 (rank : 4) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 571 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O35231, Q91YQ2 | Gene names | Kifc3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
KIFC3_HUMAN
|
||||||
| NC score | 0.060870 (rank : 5) | θ value | 0.0563607 (rank : 5) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 619 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BVG8, O75299, Q8IUT3, Q96HW6 | Gene names | KIFC3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC3. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.058925 (rank : 6) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
K1H2_HUMAN
|
||||||
| NC score | 0.052872 (rank : 7) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14532 | Gene names | KRTHA2, HHA2, HKA2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.050679 (rank : 8) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.048759 (rank : 9) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.046375 (rank : 10) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
K1H7_HUMAN
|
||||||
| NC score | 0.044928 (rank : 11) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O76014 | Gene names | KRTHA7, HHA7, HKA7 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha7 (Hair keratin, type I Ha7). | |||||
|
RPB4_HUMAN
|
||||||
| NC score | 0.044772 (rank : 12) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15514, Q52LT4 | Gene names | POLR2D | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II 16 kDa polypeptide (EC 2.7.7.6) (RPB4). | |||||
|
RPB4_MOUSE
|
||||||
| NC score | 0.044772 (rank : 13) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D7M8, Q6ZWW0, Q78KE6 | Gene names | Polr2d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA-directed RNA polymerase II 16 kDa polypeptide (EC 2.7.7.6) (RPB4). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.044722 (rank : 14) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
XRCC4_HUMAN
|
||||||
| NC score | 0.044484 (rank : 15) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13426, Q9BS72, Q9UP94 | Gene names | XRCC4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein XRCC4 (X-ray repair cross-complementing protein 4). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.044035 (rank : 16) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
K1H1_MOUSE
|
||||||
| NC score | 0.041750 (rank : 17) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61765 | Gene names | Krtha1, Hka1, Krt1-1 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha1 (Hair keratin, type I Ha1) (HKA-1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.041612 (rank : 18) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.040297 (rank : 19) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
BRE1B_HUMAN
|
||||||
| NC score | 0.039891 (rank : 20) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75150, Q6AHZ6, Q6N005, Q7L3T6, Q8N615, Q96T18, Q9BSV9, Q9HC82 | Gene names | RNF40, BRE1B, KIAA0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40) (95 kDa retinoblastoma-associated protein) (RBP95). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.039099 (rank : 21) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.038817 (rank : 22) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.038628 (rank : 23) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
MY18A_HUMAN
|
||||||
| NC score | 0.038532 (rank : 24) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1929 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92614, Q8IXP8 | Gene names | MYO18A, KIAA0216, MYSPDZ | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
BRE1B_MOUSE
|
||||||
| NC score | 0.038472 (rank : 25) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 970 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q3U319, Q6ZQ75, Q8BJA1, Q8BY03, Q8CHX4 | Gene names | Rnf40, Bre1b, Kiaa0661 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1B (EC 6.3.2.-) (BRE1-B) (RING finger protein 40). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.037905 (rank : 26) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.037538 (rank : 27) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.036673 (rank : 28) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.036584 (rank : 29) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STIM1_HUMAN
|
||||||
| NC score | 0.034367 (rank : 30) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13586 | Gene names | STIM1, GOK | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.034349 (rank : 31) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.033620 (rank : 32) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.033012 (rank : 33) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.032908 (rank : 34) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.032473 (rank : 35) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.020262 (rank : 36) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.020090 (rank : 37) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.015931 (rank : 38) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 38 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||