Please be patient as the page loads
|
GCN1L_HUMAN
|
||||||
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GCN1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
ECM29_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 2) | NC score | 0.294421 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VYK3, O15074, Q8WU82 | Gene names | ECM29, KIAA0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
ECM29_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 3) | NC score | 0.285792 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
2AAA_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 4) | NC score | 0.309525 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q76MZ3 | Gene names | Ppp2r1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform). | |||||
|
2AAA_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 5) | NC score | 0.308415 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30153, Q13773 | Gene names | PPP2R1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein). | |||||
|
2AAB_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 6) | NC score | 0.305698 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30154, O75620 | Gene names | PPP2R1B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
2AAB_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 7) | NC score | 0.297839 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNP2 | Gene names | Ppp2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
HEAT2_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 8) | NC score | 0.259549 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
IPO4_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 9) | NC score | 0.214496 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TEX9, Q86TZ9, Q8NCG8, Q96SJ3, Q9BTI4, Q9H5L0 | Gene names | IPO4, IMP4B, RANBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4). | |||||
|
IPO4_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 10) | NC score | 0.192243 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
CAND1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 11) | NC score | 0.167885 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
CAND1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 12) | NC score | 0.164898 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 13) | NC score | 0.114586 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.113080 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
CAND2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.162424 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
PLAK_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.107144 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.107065 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.068633 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
IMA4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.084247 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |
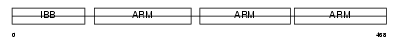
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
IMA4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 20) | NC score | 0.084320 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |
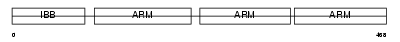
|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.140631 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 22) | NC score | 0.139018 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 23) | NC score | 0.059686 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CLAP1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.068161 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
MMS19_MOUSE
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.100517 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D071, Q925N8, Q9CWX3 | Gene names | Mms19l, Mms19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (MET18 homolog). | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 0.163984 (rank : 26) | NC score | 0.071639 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.057508 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.365318 (rank : 28) | NC score | 0.050498 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MMS19_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.093505 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T76, Q5T455, Q66K82, Q7L4W8, Q969Z1, Q96DF1, Q96MR1, Q96RK5, Q96SK1, Q9BUE2, Q9BYS9 | Gene names | MMS19L, MMS19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (hMMS19) (MET18 homolog). | |||||
|
MOT8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.041157 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
CAND2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.155618 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
COG3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.070897 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JB2, Q5VT70, Q8IXX4, Q9BZ92 | Gene names | COG3, SEC34 | |||
|
Domain Architecture |
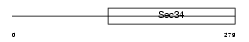
|
|||||
| Description | Conserved oligomeric Golgi complex component 3 (Vesicle docking protein SEC34 homolog) (p94). | |||||
|
TNPO1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.129814 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
TNPO1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.129637 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.049488 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.045506 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.072266 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
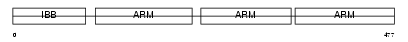
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.073272 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.054233 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RFIP3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.049740 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.039393 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
TNPO2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.119163 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.121874 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
ZFP27_MOUSE
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.000504 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10077, Q6PCZ6, Q8CED4 | Gene names | Zfp27, Mkr4, Zfp-27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 27 (Zfp-27) (Protein mKR4). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.041209 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
COG3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.063837 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CI04 | Gene names | Cog3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Conserved oligomeric Golgi complex component 3. | |||||
|
K0423_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.055842 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.042281 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CTNB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.090194 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.090198 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
HD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.061707 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
PRR8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.048340 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.041491 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HD_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.053938 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
IMB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.110166 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
IMB3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.106375 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00410, O15257 | Gene names | RANBP5, KPNB3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
IMB3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.106666 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BKC5, Q3TEG2, Q7TQC6, Q9EQ30 | Gene names | Ranbp5, Kpnb3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.041092 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.047389 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SPAG6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.076844 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
TRI25_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.019732 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
AT8A1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.015359 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
BTAF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.018256 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
CCD50_HUMAN
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.051730 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IVM0, Q86VH7 | Gene names | CCDC50, C3orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
CKAP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.049635 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.041804 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
KIF23_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.028113 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
LDH6A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.018524 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZMR3 | Gene names | LDHAL6A, LDHL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-lactate dehydrogenase A-like 6A (EC 1.1.1.27). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.040904 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PFD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.058133 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15212 | Gene names | PFDN6, HKE2, PFD6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
PFD6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.056148 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q03958 | Gene names | Pfdn6, H2-Ke2, Pfd6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.020599 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.022236 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.036263 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AT8A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.014621 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.044876 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
RASL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.012613 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.041758 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.042096 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
APOB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.034217 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04114, O00502, P78479, P78480, P78481, Q13779, Q13785, Q13786, Q13787, Q13788, Q7Z600, Q9UMN0 | Gene names | APOB | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein B-100 precursor (Apo B-100) [Contains: Apolipoprotein B-48 (Apo B-48)]. | |||||
|
CCD50_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.046303 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q810U5, Q3TRW1, Q8BP82, Q9CZT1, Q9D436 | Gene names | Ccdc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
FES_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.002462 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16879, Q62122 | Gene names | Fes, Fps | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.040370 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.039592 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.041705 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
IMB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.102910 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
MRCKB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.017618 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.040262 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SYHH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.019923 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49590 | Gene names | HARSL, HARSR, HO3 | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase homolog (EC 6.1.1.21) (Histidine--tRNA ligase homolog) (HisRS). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.009049 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
ZMYM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.021367 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.040558 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.045744 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.043761 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KI13A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.011608 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
MYO5C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.026293 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.009881 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
RNBP6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.085216 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60518, Q5T7X4, Q7Z3V2, Q96E78 | Gene names | RANBP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 6 (RanBP6). | |||||
|
STK36_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.003751 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
ULK4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.003867 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3V129, Q2VP87, Q32LZ6, Q8BLS0, Q8C8Z5, Q9D4H6 | Gene names | Ulk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK4 (EC 2.7.11.1) (Unc-51-like kinase 4). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.037719 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
B4GT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.008690 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
BFSP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.027018 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 508 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12934, O43595, O76034, Q9HBX4 | Gene names | BFSP1 | |||
|
Domain Architecture |
|
|||||
| Description | Filensin (Beaded filament structural protein 1) (Lens fiber cell beaded-filament structural protein CP 115) (CP115) (Lens intermediate filament-like heavy) (LIFL-H). | |||||
|
MACOI_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.039199 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.028367 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.028139 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
PCDGK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.001515 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.017354 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.020002 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
TEX10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.021981 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXF1, Q5T722, Q5T723, Q8NCN8, Q8TDY0 | Gene names | TEX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 10 protein. | |||||
|
TNNI3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.018324 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48787 | Gene names | Tnni3 | |||
|
Domain Architecture |
|
|||||
| Description | Troponin I, cardiac muscle (Cardiac troponin I). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.035890 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
IMA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.056067 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
IMA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.053443 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |
|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.052751 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
PP4R1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.061125 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
PP4R1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.057858 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
PP4RL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.070398 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P1A2, Q9BZ17, Q9BZ18 | Gene names | PPP4R1L, C20orf192 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1-like. | |||||
|
SPAG6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.060400 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
GCN1L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q92616, O95001, O95651, Q6P2S3, Q86X65, Q8N5I5, Q8WU80, Q99736, Q9UE60 | Gene names | GCN1L1, KIAA0219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GCN1-like protein 1 (HsGCN1). | |||||
|
2AAA_MOUSE
|
||||||
| NC score | 0.309525 (rank : 2) | θ value | 4.45536e-07 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q76MZ3 | Gene names | Ppp2r1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform). | |||||
|
2AAA_HUMAN
|
||||||
| NC score | 0.308415 (rank : 3) | θ value | 5.81887e-07 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P30153, Q13773 | Gene names | PPP2R1A | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PP2A, subunit A, PR65-alpha isoform) (PP2A, subunit A, R1-alpha isoform) (Medium tumor antigen-associated 61 kDa protein). | |||||
|
2AAB_HUMAN
|
||||||
| NC score | 0.305698 (rank : 4) | θ value | 1.69304e-06 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30154, O75620 | Gene names | PPP2R1B | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
2AAB_MOUSE
|
||||||
| NC score | 0.297839 (rank : 5) | θ value | 8.40245e-06 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TNP2 | Gene names | Ppp2r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform (PP2A, subunit A, PR65-beta isoform) (PP2A, subunit A, R1-beta isoform). | |||||
|
ECM29_HUMAN
|
||||||
| NC score | 0.294421 (rank : 6) | θ value | 1.38499e-08 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5VYK3, O15074, Q8WU82 | Gene names | ECM29, KIAA0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
ECM29_MOUSE
|
||||||
| NC score | 0.285792 (rank : 7) | θ value | 5.26297e-08 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PDI5, Q6ZQC9, Q8BSW7, Q8CAH0, Q8R3M6 | Gene names | Ecm29, Kiaa0368 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteasome-associated protein ECM29 homolog (Ecm29). | |||||
|
HEAT2_HUMAN
|
||||||
| NC score | 0.259549 (rank : 8) | θ value | 9.29e-05 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86Y56, Q69YL1, Q96FI9, Q9NX75 | Gene names | HEATR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 2. | |||||
|
IPO4_HUMAN
|
||||||
| NC score | 0.214496 (rank : 9) | θ value | 0.000158464 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TEX9, Q86TZ9, Q8NCG8, Q96SJ3, Q9BTI4, Q9H5L0 | Gene names | IPO4, IMP4B, RANBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-4 (Importin 4b) (Imp4b) (Ran-binding protein 4) (RanBP4). | |||||
|
IPO4_MOUSE
|
||||||
| NC score | 0.192243 (rank : 10) | θ value | 0.00298849 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VI75, Q3TBW0, Q99J52 | Gene names | Ipo4, Imp4a, Ranbp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Importin-4 (Importin 4a) (Imp4a) (Ran-binding protein 4) (RanBP4). | |||||
|
CAND1_HUMAN
|
||||||
| NC score | 0.167885 (rank : 11) | θ value | 0.00390308 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q86VP6, O94918, Q6PIY4, Q8NDJ4, Q96JZ9, Q96T19, Q9BTC4, Q9H0G2, Q9P0H7, Q9UF85 | Gene names | CAND1, KIAA0829, TIP120, TIP120A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1) (TBP-interacting protein TIP120A) (TBP-interacting protein of 120 kDa A). | |||||
|
CAND1_MOUSE
|
||||||
| NC score | 0.164898 (rank : 12) | θ value | 0.00509761 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZQ38, Q6PFR0, Q9CV45 | Gene names | Cand1, D10Ertd516e, Kiaa0829 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 1 (Cullin-associated and neddylation-dissociated protein 1) (p120 CAND1). | |||||
|
CAND2_MOUSE
|
||||||
| NC score | 0.162424 (rank : 13) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZQ73, Q6PHT7 | Gene names | Cand2, Kiaa0667, Tip120b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2) (TBP-interacting protein TIP120B) (TBP-interacting protein of 120 kDa B). | |||||
|
CAND2_HUMAN
|
||||||
| NC score | 0.155618 (rank : 14) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75155 | Gene names | CAND2, KIAA0667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cullin-associated NEDD8-dissociated protein 2 (Cullin-associated and neddylation-dissociated protein 2) (p120 CAND2). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.140631 (rank : 15) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.139018 (rank : 16) | θ value | 0.0563607 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
TNPO1_HUMAN
|
||||||
| NC score | 0.129814 (rank : 17) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92973, Q92957, Q92975 | Gene names | TNPO1, KPNB2, MIP1, TRN | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2) (M9 region interaction protein) (MIP). | |||||
|
TNPO1_MOUSE
|
||||||
| NC score | 0.129637 (rank : 18) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BFY9, Q8C0S3, Q8K2E7 | Gene names | Tnpo1, Kpnb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-1 (Importin beta-2) (Karyopherin beta-2). | |||||
|
TNPO2_MOUSE
|
||||||
| NC score | 0.121874 (rank : 19) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99LG2, Q8C0U9 | Gene names | Tnpo2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
TNPO2_HUMAN
|
||||||
| NC score | 0.119163 (rank : 20) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14787, O14655 | Gene names | TNPO2 | |||
|
Domain Architecture |

|
|||||
| Description | Transportin-2 (Karyopherin beta-2b). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.114586 (rank : 21) | θ value | 0.00869519 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.113080 (rank : 22) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
IMB1_MOUSE
|
||||||
| NC score | 0.110166 (rank : 23) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P70168, Q62117 | Gene names | Kpnb1, Impnb | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Pore targeting complex 97 kDa subunit) (PTAC97) (SCG). | |||||
|
PLAK_MOUSE
|
||||||
| NC score | 0.107144 (rank : 24) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02257, Q8CGD3 | Gene names | Jup | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
PLAK_HUMAN
|
||||||
| NC score | 0.107065 (rank : 25) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P14923, Q9HCX9 | Gene names | JUP, DP3 | |||
|
Domain Architecture |

|
|||||
| Description | Junction plakoglobin (Desmoplakin-3) (Desmoplakin III). | |||||
|
IMB3_MOUSE
|
||||||
| NC score | 0.106666 (rank : 26) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BKC5, Q3TEG2, Q7TQC6, Q9EQ30 | Gene names | Ranbp5, Kpnb3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
IMB3_HUMAN
|
||||||
| NC score | 0.106375 (rank : 27) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00410, O15257 | Gene names | RANBP5, KPNB3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-3 (Karyopherin beta-3) (Ran-binding protein 5) (RanBP5). | |||||
|
IMB1_HUMAN
|
||||||
| NC score | 0.102910 (rank : 28) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14974, Q14637, Q96J27 | Gene names | KPNB1, NTF97 | |||
|
Domain Architecture |

|
|||||
| Description | Importin beta-1 subunit (Karyopherin beta-1 subunit) (Nuclear factor P97) (Importin 90). | |||||
|
MMS19_MOUSE
|
||||||
| NC score | 0.100517 (rank : 29) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D071, Q925N8, Q9CWX3 | Gene names | Mms19l, Mms19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (MET18 homolog). | |||||
|
MMS19_HUMAN
|
||||||
| NC score | 0.093505 (rank : 30) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96T76, Q5T455, Q66K82, Q7L4W8, Q969Z1, Q96DF1, Q96MR1, Q96RK5, Q96SK1, Q9BUE2, Q9BYS9 | Gene names | MMS19L, MMS19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MMS19-like protein (hMMS19) (MET18 homolog). | |||||
|
CTNB1_MOUSE
|
||||||
| NC score | 0.090198 (rank : 31) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02248, Q922W1, Q9D335 | Gene names | Ctnnb1, Catnb | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
CTNB1_HUMAN
|
||||||
| NC score | 0.090194 (rank : 32) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P35222, Q8NEW9, Q8NI94, Q9H391 | Gene names | CTNNB1, CTNNB | |||
|
Domain Architecture |

|
|||||
| Description | Catenin beta-1 (Beta-catenin). | |||||
|
RNBP6_HUMAN
|
||||||
| NC score | 0.085216 (rank : 33) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60518, Q5T7X4, Q7Z3V2, Q96E78 | Gene names | RANBP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 6 (RanBP6). | |||||
|
IMA4_MOUSE
|
||||||
| NC score | 0.084320 (rank : 34) | θ value | 0.0563607 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35343 | Gene names | Kpna4 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Importin alpha Q1). | |||||
|
IMA4_HUMAN
|
||||||
| NC score | 0.084247 (rank : 35) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00629, O00190 | Gene names | KPNA4, QIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-4 subunit (Karyopherin alpha-4 subunit) (Qip1 protein). | |||||
|
SPAG6_MOUSE
|
||||||
| NC score | 0.076844 (rank : 36) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9JLI7, Q8K461 | Gene names | Spag6, Pf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Axoneme central apparatus protein). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.073272 (rank : 37) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.072266 (rank : 38) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.071639 (rank : 39) | θ value | 0.163984 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
COG3_HUMAN
|
||||||
| NC score | 0.070897 (rank : 40) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JB2, Q5VT70, Q8IXX4, Q9BZ92 | Gene names | COG3, SEC34 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 3 (Vesicle docking protein SEC34 homolog) (p94). | |||||
|
PP4RL_HUMAN
|
||||||
| NC score | 0.070398 (rank : 41) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P1A2, Q9BZ17, Q9BZ18 | Gene names | PPP4R1L, C20orf192 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1-like. | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.068633 (rank : 42) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CLAP1_HUMAN
|
||||||
| NC score | 0.068161 (rank : 43) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z460, O75118, Q8N5B8, Q9BQT5 | Gene names | CLASP1, KIAA0622, MAST1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 1 (Cytoplasmic linker-associated protein 1) (Multiple asters homolog 1). | |||||
|
COG3_MOUSE
|
||||||
| NC score | 0.063837 (rank : 44) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CI04 | Gene names | Cog3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Conserved oligomeric Golgi complex component 3. | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.061707 (rank : 45) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
PP4R1_HUMAN
|
||||||
| NC score | 0.061125 (rank : 46) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
SPAG6_HUMAN
|
||||||
| NC score | 0.060400 (rank : 47) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75602, Q5VUX5, Q5VUX6, Q5VUX7, Q6FI74, Q8NHQ6 | Gene names | SPAG6, PF16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 6 (PF16 protein homolog) (Sperm flagellar protein) (Repro-SA-1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.059686 (rank : 48) | θ value | 0.0736092 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
PFD6_HUMAN
|
||||||
| NC score | 0.058133 (rank : 49) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15212 | Gene names | PFDN6, HKE2, PFD6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
PP4R1_MOUSE
|
||||||
| NC score | 0.057858 (rank : 50) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2V1, Q8R0R8 | Gene names | Ppp4r1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.057508 (rank : 51) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
PFD6_MOUSE
|
||||||
| NC score | 0.056148 (rank : 52) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q03958 | Gene names | Pfdn6, H2-Ke2, Pfd6 | |||
|
Domain Architecture |

|
|||||
| Description | Prefoldin subunit 6 (Protein Ke2). | |||||
|
IMA2_HUMAN
|
||||||
| NC score | 0.056067 (rank : 53) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52292, Q9BRU5 | Gene names | KPNA2, RCH1, SRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1). | |||||
|
K0423_HUMAN
|
||||||
| NC score | 0.055842 (rank : 54) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y4F4, Q68D66, Q6PG27 | Gene names | KIAA0423 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0423. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.054233 (rank : 55) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.053938 (rank : 56) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
IMA2_MOUSE
|
||||||
| NC score | 0.053443 (rank : 57) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52293, Q64292 | Gene names | Kpna2, Rch1 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-2 subunit (Karyopherin alpha-2 subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin) (Pore targeting complex 58 kDa subunit) (PTAC58) (Importin alpha P1). | |||||
|
IMA5_HUMAN
|
||||||
| NC score | 0.052751 (rank : 58) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15131 | Gene names | KPNA5 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-6 subunit (Karyopherin alpha-5 subunit). | |||||
|
CCD50_HUMAN
|
||||||
| NC score | 0.051730 (rank : 59) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8IVM0, Q86VH7 | Gene names | CCDC50, C3orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.050498 (rank : 60) | θ value | 0.365318 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
RFIP3_HUMAN
|
||||||
| NC score | 0.049740 (rank : 61) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1055 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O75154, Q4VXV7, Q9H155, Q9H1G0, Q9NUI0 | Gene names | RAB11FIP3, KIAA0665 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 3 (Rab11-FIP3) (EF hands-containing Rab-interacting protein) (Eferin). | |||||
|
CKAP5_HUMAN
|
||||||
| NC score | 0.049635 (rank : 62) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14008, Q14668 | Gene names | CKAP5, KIAA0097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 5 (Colonic and hepatic tumor over- expressed protein) (Ch-TOG protein). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.049488 (rank : 63) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.048340 (rank : 64) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.047389 (rank : 65) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
CCD50_MOUSE
|
||||||
| NC score | 0.046303 (rank : 66) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q810U5, Q3TRW1, Q8BP82, Q9CZT1, Q9D436 | Gene names | Ccdc50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 50 (Protein Ymer). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.045744 (rank : 67) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.045506 (rank : 68) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.044876 (rank : 69) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.043761 (rank : 70) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.042281 (rank : 71) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.042096 (rank : 72) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.041804 (rank : 73) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.041758 (rank : 74) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.041705 (rank : 75) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.041491 (rank : 76) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.041209 (rank : 77) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MOT8_HUMAN
|
||||||
| NC score | 0.041157 (rank : 78) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P36021, Q7Z797 | Gene names | SLC16A2, MCT8, XPCT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Monocarboxylate transporter 8 (MCT 8) (MCT 7) (Solute carrier family 16 member 2) (X-linked PEST-containing transporter). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.041092 (rank : 79) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.040904 (rank : 80) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.040558 (rank : 81) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.040370 (rank : 82) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.040262 (rank : 83) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.039592 (rank : 84) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.039393 (rank : 85) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MACOI_HUMAN
|
||||||
| NC score | 0.039199 (rank : 86) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N5G2, Q2TLX5, Q2TLX6, Q9NVG6 | Gene names | TMEM57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Macoilin (Transmembrane protein 57). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.037719 (rank : 87) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.036263 (rank : 88) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.035890 (rank : 89) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
APOB_HUMAN
|
||||||
| NC score | 0.034217 (rank : 90) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04114, O00502, P78479, P78480, P78481, Q13779, Q13785, Q13786, Q13787, Q13788, Q7Z600, Q9UMN0 | Gene names | APOB | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein B-100 precursor (Apo B-100) [Contains: Apolipoprotein B-48 (Apo B-48)]. | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.028367 (rank : 91) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.028139 (rank : 92) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
KIF23_HUMAN
|
||||||
| NC score | 0.028113 (rank : 93) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02241 | Gene names | KIF23, KNSL5, MKLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF23 (Mitotic kinesin-like protein 1) (Kinesin- like protein 5). | |||||
|
BFSP1_HUMAN
|
||||||
| NC score | 0.027018 (rank : 94) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 508 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q12934, O43595, O76034, Q9HBX4 | Gene names | BFSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filensin (Beaded filament structural protein 1) (Lens fiber cell beaded-filament structural protein CP 115) (CP115) (Lens intermediate filament-like heavy) (LIFL-H). | |||||
|
MYO5C_HUMAN
|
||||||
| NC score | 0.026293 (rank : 95) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1206 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NQX4 | Gene names | MYO5C | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5C (Myosin Vc). | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.022236 (rank : 96) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
TEX10_HUMAN
|
||||||
| NC score | 0.021981 (rank : 97) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NXF1, Q5T722, Q5T723, Q8NCN8, Q8TDY0 | Gene names | TEX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 10 protein. | |||||
|
ZMYM1_HUMAN
|
||||||
| NC score | 0.021367 (rank : 98) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5SVZ6, Q7Z3Q4 | Gene names | ZMYM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 1. | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.020599 (rank : 99) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.020002 (rank : 100) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
SYHH_HUMAN
|
||||||
| NC score | 0.019923 (rank : 101) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49590 | Gene names | HARSL, HARSR, HO3 | |||
|
Domain Architecture |

|
|||||
| Description | Histidyl-tRNA synthetase homolog (EC 6.1.1.21) (Histidine--tRNA ligase homolog) (HisRS). | |||||
|
TRI25_HUMAN
|
||||||
| NC score | 0.019732 (rank : 102) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
LDH6A_HUMAN
|
||||||
| NC score | 0.018524 (rank : 103) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZMR3 | Gene names | LDHAL6A, LDHL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | L-lactate dehydrogenase A-like 6A (EC 1.1.1.27). | |||||
|
TNNI3_MOUSE
|
||||||
| NC score | 0.018324 (rank : 104) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48787 | Gene names | Tnni3 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin I, cardiac muscle (Cardiac troponin I). | |||||
|
BTAF1_HUMAN
|
||||||
| NC score | 0.018256 (rank : 105) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O14981, O43578 | Gene names | BTAF1, TAF172 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 172 (EC 3.6.1.-) (ATP-dependent helicase BTAF1) (TBP-associated factor 172) (TAF-172) (TAF(II)170) (B- TFIID transcription factor-associated 170 kDa subunit). | |||||
|
MRCKB_MOUSE
|
||||||
| NC score | 0.017618 (rank : 106) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2180 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q7TT50, Q69ZR5, Q80W33 | Gene names | Cdc42bpb, Kiaa1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.017354 (rank : 107) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
AT8A1_HUMAN
|
||||||
| NC score | 0.015359 (rank : 108) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2Q0 | Gene names | ATP8A1, ATPIA | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
AT8A1_MOUSE
|
||||||
| NC score | 0.014621 (rank : 109) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70704 | Gene names | Atp8a1, Atpc1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IA (EC 3.6.3.1) (Chromaffin granule ATPase II) (ATPase class I type 8A member 1). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.012613 (rank : 110) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
KI13A_MOUSE
|
||||||
| NC score | 0.011608 (rank : 111) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.009881 (rank : 112) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.009049 (rank : 113) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
B4GT2_MOUSE
|
||||||
| NC score | 0.008690 (rank : 114) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2Y2, Q3TMP2 | Gene names | B4galt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-1,4-galactosyltransferase 2 (EC 2.4.1.-) (Beta-1,4-GalTase 2) (Beta4Gal-T2) (b4Gal-T2) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 2) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 2) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
ULK4_MOUSE
|
||||||
| NC score | 0.003867 (rank : 115) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3V129, Q2VP87, Q32LZ6, Q8BLS0, Q8C8Z5, Q9D4H6 | Gene names | Ulk4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK4 (EC 2.7.11.1) (Unc-51-like kinase 4). | |||||
|
STK36_HUMAN
|
||||||
| NC score | 0.003751 (rank : 116) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRP7, Q8TC32, Q9H9N9, Q9UF35, Q9ULE2 | Gene names | STK36, KIAA1278 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase 36 (EC 2.7.11.1) (Fused homolog). | |||||
|
FES_MOUSE
|
||||||
| NC score | 0.002462 (rank : 117) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 980 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16879, Q62122 | Gene names | Fes, Fps | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase Fes/Fps (EC 2.7.10.2) (C-Fes). | |||||
|
PCDGK_HUMAN
|
||||||
| NC score | 0.001515 (rank : 118) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
ZFP27_MOUSE
|
||||||
| NC score | 0.000504 (rank : 119) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10077, Q6PCZ6, Q8CED4 | Gene names | Zfp27, Mkr4, Zfp-27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 27 (Zfp-27) (Protein mKR4). | |||||