Please be patient as the page loads
|
CDY2_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CDY1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999771 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987247 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.990115 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 1.53757e-108 (rank : 5) | NC score | 0.975639 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
PECI_MOUSE
|
||||||
| θ value | 4.25575e-50 (rank : 6) | NC score | 0.816034 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
PECI_HUMAN
|
||||||
| θ value | 2.18568e-46 (rank : 7) | NC score | 0.802926 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 8) | NC score | 0.674353 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 9) | NC score | 0.670177 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECH1_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 10) | NC score | 0.674440 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 11) | NC score | 0.679768 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 12) | NC score | 0.212922 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
AUMH_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 13) | NC score | 0.574542 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
AUHM_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 14) | NC score | 0.579987 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
ECHP_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 15) | NC score | 0.439709 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
ECHA_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 16) | NC score | 0.464938 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 17) | NC score | 0.468515 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 7.59969e-07 (rank : 18) | NC score | 0.457600 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
ECHP_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 19) | NC score | 0.393936 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 20) | NC score | 0.456279 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 21) | NC score | 0.456279 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 22) | NC score | 0.455612 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 23) | NC score | 0.454635 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 24) | NC score | 0.416319 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 25) | NC score | 0.433958 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 26) | NC score | 0.354893 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 27) | NC score | 0.384039 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 28) | NC score | 0.366896 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 29) | NC score | 0.365787 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 30) | NC score | 0.373975 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 31) | NC score | 0.376211 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 32) | NC score | 0.366627 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 33) | NC score | 0.370803 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 34) | NC score | 0.212225 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 35) | NC score | 0.221594 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 36) | NC score | 0.205228 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 37) | NC score | 0.205157 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 38) | NC score | 0.088046 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 39) | NC score | 0.084236 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 40) | NC score | 0.109311 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 41) | NC score | 0.060025 (rank : 52) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.086096 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.060845 (rank : 51) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.007829 (rank : 57) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
SIX1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.004544 (rank : 58) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15475, Q96H64 | Gene names | SIX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX1 (Sine oculis homeobox homolog 1). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.043377 (rank : 55) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.044804 (rank : 53) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.044413 (rank : 54) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.008983 (rank : 56) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
ACBD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.108864 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC06, Q8IUT1, Q9H8Q4 | Gene names | ACBD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 4. | |||||
|
ACBD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.114281 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6N7 | Gene names | ACBD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 7. | |||||
|
ACBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.119761 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07108, P08869, Q53SQ7, Q6IB48 | Gene names | DBI | |||
|
Domain Architecture |
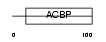
|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
ACBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.121465 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31786 | Gene names | Dbi | |||
|
Domain Architecture |
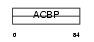
|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
D3D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.194422 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42126, Q13290, Q7Z2L6, Q9BUB8, Q9BW05 | Gene names | DCI | |||
|
Domain Architecture |
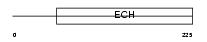
|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
D3D2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.177167 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42125 | Gene names | Dci | |||
|
Domain Architecture |
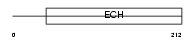
|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
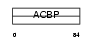
DBIL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.111828 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09035 | Gene names | Dbil5 | |||
|
Domain Architecture |
|
|||||
| Description | Diazepam-binding inhibitor-like 5 (Endozepine-like peptide) (ELP). | |||||
|
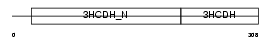
HCDH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.073237 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
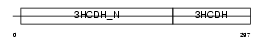
HCDH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.072061 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.999771 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.990115 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.987247 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.975639 (rank : 5) | θ value | 1.53757e-108 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.816034 (rank : 6) | θ value | 4.25575e-50 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.802926 (rank : 7) | θ value | 2.18568e-46 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.679768 (rank : 8) | θ value | 1.29331e-14 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.674440 (rank : 9) | θ value | 4.44505e-15 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.674353 (rank : 10) | θ value | 2.5182e-18 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.670177 (rank : 11) | θ value | 3.63628e-17 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.579987 (rank : 12) | θ value | 1.63604e-09 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.574542 (rank : 13) | θ value | 4.30538e-10 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.468515 (rank : 14) | θ value | 2.61198e-07 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
ECHA_HUMAN
|
||||||
| NC score | 0.464938 (rank : 15) | θ value | 5.26297e-08 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.457600 (rank : 16) | θ value | 7.59969e-07 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.456279 (rank : 17) | θ value | 2.21117e-06 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.456279 (rank : 18) | θ value | 2.21117e-06 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.455612 (rank : 19) | θ value | 2.21117e-06 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.454635 (rank : 20) | θ value | 2.21117e-06 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
ECHP_MOUSE
|
||||||
| NC score | 0.439709 (rank : 21) | θ value | 1.06045e-08 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.433958 (rank : 22) | θ value | 4.1701e-05 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.416319 (rank : 23) | θ value | 4.1701e-05 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
ECHP_HUMAN
|
||||||
| NC score | 0.393936 (rank : 24) | θ value | 7.59969e-07 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.384039 (rank : 25) | θ value | 0.000121331 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.376211 (rank : 26) | θ value | 0.000121331 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.373975 (rank : 27) | θ value | 0.000121331 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.370803 (rank : 28) | θ value | 0.00020696 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.366896 (rank : 29) | θ value | 0.000121331 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.366627 (rank : 30) | θ value | 0.00020696 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.365787 (rank : 31) | θ value | 0.000121331 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.354893 (rank : 32) | θ value | 0.000121331 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.221594 (rank : 33) | θ value | 0.000786445 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.212922 (rank : 34) | θ value | 1.133e-10 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.212225 (rank : 35) | θ value | 0.000786445 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.205228 (rank : 36) | θ value | 0.00509761 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.205157 (rank : 37) | θ value | 0.0330416 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
D3D2_HUMAN
|
||||||
| NC score | 0.194422 (rank : 38) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42126, Q13290, Q7Z2L6, Q9BUB8, Q9BW05 | Gene names | DCI | |||
|
Domain Architecture |

|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
D3D2_MOUSE
|
||||||
| NC score | 0.177167 (rank : 39) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42125 | Gene names | Dci | |||
|
Domain Architecture |

|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
ACBP_MOUSE
|
||||||
| NC score | 0.121465 (rank : 40) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31786 | Gene names | Dbi | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
ACBP_HUMAN
|
||||||
| NC score | 0.119761 (rank : 41) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07108, P08869, Q53SQ7, Q6IB48 | Gene names | DBI | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
ACBD7_HUMAN
|
||||||
| NC score | 0.114281 (rank : 42) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6N7 | Gene names | ACBD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 7. | |||||
|
DBIL5_MOUSE
|
||||||
| NC score | 0.111828 (rank : 43) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09035 | Gene names | Dbil5 | |||
|
Domain Architecture |

|
|||||
| Description | Diazepam-binding inhibitor-like 5 (Endozepine-like peptide) (ELP). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.109311 (rank : 44) | θ value | 0.279714 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
ACBD4_HUMAN
|
||||||
| NC score | 0.108864 (rank : 45) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC06, Q8IUT1, Q9H8Q4 | Gene names | ACBD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 4. | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.088046 (rank : 46) | θ value | 0.163984 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.086096 (rank : 47) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.084236 (rank : 48) | θ value | 0.279714 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
HCDH_HUMAN
|
||||||
| NC score | 0.073237 (rank : 49) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
HCDH_MOUSE
|
||||||
| NC score | 0.072061 (rank : 50) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.060845 (rank : 51) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.060025 (rank : 52) | θ value | 0.365318 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.044804 (rank : 53) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.044413 (rank : 54) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.043377 (rank : 55) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.008983 (rank : 56) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.007829 (rank : 57) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
SIX1_HUMAN
|
||||||
| NC score | 0.004544 (rank : 58) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15475, Q96H64 | Gene names | SIX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX1 (Sine oculis homeobox homolog 1). | |||||