Please be patient as the page loads
|
CDYL1_HUMAN
|
||||||
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CDY1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987248 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987247 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.992025 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 7.32286e-135 (rank : 5) | NC score | 0.976856 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
PECI_MOUSE
|
||||||
| θ value | 3.04986e-48 (rank : 6) | NC score | 0.799938 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
PECI_HUMAN
|
||||||
| θ value | 6.7944e-48 (rank : 7) | NC score | 0.786637 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 8) | NC score | 0.668924 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECH1_MOUSE
|
||||||
| θ value | 9.56915e-18 (rank : 9) | NC score | 0.681751 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 10) | NC score | 0.676033 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 11) | NC score | 0.664719 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 12) | NC score | 0.224548 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
AUMH_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 13) | NC score | 0.576310 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
AUHM_MOUSE
|
||||||
| θ value | 5.08577e-11 (rank : 14) | NC score | 0.581623 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
ECHA_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 15) | NC score | 0.474191 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 3.29651e-10 (rank : 16) | NC score | 0.493724 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 17) | NC score | 0.503697 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
ECHP_MOUSE
|
||||||
| θ value | 5.62301e-10 (rank : 18) | NC score | 0.444689 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 19) | NC score | 0.495497 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 20) | NC score | 0.491546 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 21) | NC score | 0.491348 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 22) | NC score | 0.491348 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
ECHP_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 23) | NC score | 0.399154 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 24) | NC score | 0.413664 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 25) | NC score | 0.382077 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 26) | NC score | 0.436922 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 27) | NC score | 0.385269 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 28) | NC score | 0.454166 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 29) | NC score | 0.383927 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 30) | NC score | 0.390798 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 31) | NC score | 0.391794 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 32) | NC score | 0.382632 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 33) | NC score | 0.386377 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 34) | NC score | 0.219226 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 35) | NC score | 0.114006 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 36) | NC score | 0.208670 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 37) | NC score | 0.090123 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 38) | NC score | 0.037185 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 39) | NC score | 0.026696 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 40) | NC score | 0.204932 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 0.125558 (rank : 41) | NC score | 0.203173 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 42) | NC score | 0.029945 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 43) | NC score | 0.114825 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.21417 (rank : 44) | NC score | 0.075354 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.21417 (rank : 45) | NC score | 0.074898 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 46) | NC score | 0.073321 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 47) | NC score | 0.034420 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 48) | NC score | 0.116106 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 49) | NC score | 0.116409 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MS3L1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 50) | NC score | 0.067541 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 0.62314 (rank : 51) | NC score | 0.049955 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
NKTR_HUMAN
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.017883 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.055504 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.031351 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.035116 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
APOA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.003397 (rank : 98) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
BRDT_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.010148 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.035427 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
SORC3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.011008 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.018863 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
TDRD5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.020441 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |
|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
GCP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.018938 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
MEF2B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.014668 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.014082 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
CACB2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.008625 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.011898 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
KLF14_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | -0.001999 (rank : 101) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.018177 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.009628 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.016599 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.016599 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.009601 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
ZN433_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | -0.002441 (rank : 102) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N7K0 | Gene names | ZNF433 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 433. | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.020176 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.054532 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
LMD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.021222 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.006298 (rank : 94) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.013570 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.006608 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
ZRAB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.019352 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
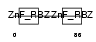
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.044673 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
FOXD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.002932 (rank : 100) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |
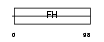
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.008038 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PACN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.003176 (rank : 99) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVE8 | Gene names | Pacsin2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
DMXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.003971 (rank : 96) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
IQEC3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.003463 (rank : 97) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.009013 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.022119 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.017015 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
TF3C2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.007202 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
VMDL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.005077 (rank : 95) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
ZN148_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | -0.003033 (rank : 103) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
ACBD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.104339 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC06, Q8IUT1, Q9H8Q4 | Gene names | ACBD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 4. | |||||
|
ACBD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.109446 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6N7 | Gene names | ACBD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 7. | |||||
|
ACBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.114661 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07108, P08869, Q53SQ7, Q6IB48 | Gene names | DBI | |||
|
Domain Architecture |
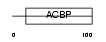
|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
ACBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.116356 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31786 | Gene names | Dbi | |||
|
Domain Architecture |
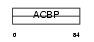
|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.051792 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051125 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
D3D2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.198802 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42126, Q13290, Q7Z2L6, Q9BUB8, Q9BW05 | Gene names | DCI | |||
|
Domain Architecture |
|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
D3D2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.180660 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42125 | Gene names | Dci | |||
|
Domain Architecture |
|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
DBIL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.107128 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09035 | Gene names | Dbil5 | |||
|
Domain Architecture |
|
|||||
| Description | Diazepam-binding inhibitor-like 5 (Endozepine-like peptide) (ELP). | |||||
|
HCDH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.080389 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
HCDH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.079090 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
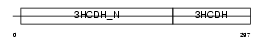
Domain Architecture |
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.992025 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.987248 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.987247 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.976856 (rank : 5) | θ value | 7.32286e-135 (rank : 5) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.799938 (rank : 6) | θ value | 3.04986e-48 (rank : 6) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.786637 (rank : 7) | θ value | 6.7944e-48 (rank : 7) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.681751 (rank : 8) | θ value | 9.56915e-18 (rank : 9) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.676033 (rank : 9) | θ value | 2.7842e-17 (rank : 10) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.668924 (rank : 10) | θ value | 3.28887e-18 (rank : 8) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.664719 (rank : 11) | θ value | 3.63628e-17 (rank : 11) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.581623 (rank : 12) | θ value | 5.08577e-11 (rank : 14) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.576310 (rank : 13) | θ value | 3.89403e-11 (rank : 13) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.503697 (rank : 14) | θ value | 4.30538e-10 (rank : 17) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.495497 (rank : 15) | θ value | 7.34386e-10 (rank : 19) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.493724 (rank : 16) | θ value | 3.29651e-10 (rank : 16) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.491546 (rank : 17) | θ value | 3.64472e-09 (rank : 20) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.491348 (rank : 18) | θ value | 1.80886e-08 (rank : 21) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.491348 (rank : 19) | θ value | 1.80886e-08 (rank : 22) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
ECHA_HUMAN
|
||||||
| NC score | 0.474191 (rank : 20) | θ value | 6.64225e-11 (rank : 15) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.454166 (rank : 21) | θ value | 1.87187e-05 (rank : 28) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
ECHP_MOUSE
|
||||||
| NC score | 0.444689 (rank : 22) | θ value | 5.62301e-10 (rank : 18) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.436922 (rank : 23) | θ value | 8.40245e-06 (rank : 26) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.413664 (rank : 24) | θ value | 6.87365e-08 (rank : 24) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
ECHP_HUMAN
|
||||||
| NC score | 0.399154 (rank : 25) | θ value | 5.26297e-08 (rank : 23) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.391794 (rank : 26) | θ value | 9.29e-05 (rank : 31) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.390798 (rank : 27) | θ value | 5.44631e-05 (rank : 30) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.386377 (rank : 28) | θ value | 0.000121331 (rank : 33) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.385269 (rank : 29) | θ value | 1.87187e-05 (rank : 27) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.383927 (rank : 30) | θ value | 2.44474e-05 (rank : 29) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.382632 (rank : 31) | θ value | 0.000121331 (rank : 32) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.382077 (rank : 32) | θ value | 2.88788e-06 (rank : 25) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.224548 (rank : 33) | θ value | 7.09661e-13 (rank : 12) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.219226 (rank : 34) | θ value | 0.00134147 (rank : 34) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.208670 (rank : 35) | θ value | 0.00509761 (rank : 36) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.204932 (rank : 36) | θ value | 0.0330416 (rank : 40) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.203173 (rank : 37) | θ value | 0.125558 (rank : 41) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
D3D2_HUMAN
|
||||||
| NC score | 0.198802 (rank : 38) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42126, Q13290, Q7Z2L6, Q9BUB8, Q9BW05 | Gene names | DCI | |||
|
Domain Architecture |

|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
D3D2_MOUSE
|
||||||
| NC score | 0.180660 (rank : 39) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P42125 | Gene names | Dci | |||
|
Domain Architecture |

|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.116409 (rank : 40) | θ value | 0.279714 (rank : 49) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
ACBP_MOUSE
|
||||||
| NC score | 0.116356 (rank : 41) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31786 | Gene names | Dbi | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.116106 (rank : 42) | θ value | 0.21417 (rank : 48) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.114825 (rank : 43) | θ value | 0.21417 (rank : 43) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
ACBP_HUMAN
|
||||||
| NC score | 0.114661 (rank : 44) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07108, P08869, Q53SQ7, Q6IB48 | Gene names | DBI | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-CoA-binding protein (ACBP) (Diazepam-binding inhibitor) (DBI) (Endozepine) (EP). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.114006 (rank : 45) | θ value | 0.00298849 (rank : 35) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
ACBD7_HUMAN
|
||||||
| NC score | 0.109446 (rank : 46) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N6N7 | Gene names | ACBD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 7. | |||||
|
DBIL5_MOUSE
|
||||||
| NC score | 0.107128 (rank : 47) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O09035 | Gene names | Dbil5 | |||
|
Domain Architecture |

|
|||||
| Description | Diazepam-binding inhibitor-like 5 (Endozepine-like peptide) (ELP). | |||||
|
ACBD4_HUMAN
|
||||||
| NC score | 0.104339 (rank : 48) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NC06, Q8IUT1, Q9H8Q4 | Gene names | ACBD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyl-CoA-binding domain-containing protein 4. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.090123 (rank : 49) | θ value | 0.0148317 (rank : 37) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
HCDH_HUMAN
|
||||||
| NC score | 0.080389 (rank : 50) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
HCDH_MOUSE
|
||||||
| NC score | 0.079090 (rank : 51) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.075354 (rank : 52) | θ value | 0.21417 (rank : 44) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.074898 (rank : 53) | θ value | 0.21417 (rank : 45) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.073321 (rank : 54) | θ value | 0.21417 (rank : 46) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
MS3L1_HUMAN
|
||||||
| NC score | 0.067541 (rank : 55) | θ value | 0.47712 (rank : 50) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N5Y2, Q9UG70, Q9Y5Z8 | Gene names | MSL3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.055504 (rank : 56) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.054532 (rank : 57) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.051792 (rank : 58) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.051125 (rank : 59) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.049955 (rank : 60) | θ value | 0.62314 (rank : 51) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.044673 (rank : 61) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.037185 (rank : 62) | θ value | 0.0252991 (rank : 38) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.035427 (rank : 63) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.035116 (rank : 64) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.034420 (rank : 65) | θ value | 0.21417 (rank : 47) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.031351 (rank : 66) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.029945 (rank : 67) | θ value | 0.21417 (rank : 42) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.026696 (rank : 68) | θ value | 0.0330416 (rank : 39) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.022119 (rank : 69) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
LMD1_HUMAN
|
||||||
| NC score | 0.021222 (rank : 70) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29536 | Gene names | LMOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Leiomodin-1 (Leiomodin, muscle form) (64 kDa autoantigen D1) (64 kDa autoantigen 1D) (64 kDa autoantigen 1D3) (Thyroid-associated ophthalmopathy autoantigen) (Smooth muscle leiomodin) (SM-Lmod). | |||||
|
TDRD5_HUMAN
|
||||||
| NC score | 0.020441 (rank : 71) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NAT2, Q5VTV0 | Gene names | TDRD5 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 5. | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.020176 (rank : 72) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
ZRAB2_HUMAN
|
||||||
| NC score | 0.019352 (rank : 73) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95218, Q9UP63 | Gene names | ZRANB2, ZIS, ZNF265 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 2 Zinc finger protein 265 (Zinc finger, splicing). | |||||
|
GCP5_HUMAN
|
||||||
| NC score | 0.018938 (rank : 74) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RT8, Q96PY8 | Gene names | TUBGCP5, GCP5, KIAA1899 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-tubulin complex component 5 (GCP-5). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.018863 (rank : 75) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.018177 (rank : 76) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
NKTR_HUMAN
|
||||||
| NC score | 0.017883 (rank : 77) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P30414 | Gene names | NKTR | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.017015 (rank : 78) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.016599 (rank : 79) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.016599 (rank : 80) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
MEF2B_MOUSE
|
||||||
| NC score | 0.014668 (rank : 81) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O55087, O55088, O55089, O55090, O55231, Q61843 | Gene names | Mef2b | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B. | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.014082 (rank : 82) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.013570 (rank : 83) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.011898 (rank : 84) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
SORC3_MOUSE
|
||||||
| NC score | 0.011008 (rank : 85) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VI51, Q9CTR4 | Gene names | Sorcs3 | |||
|
Domain Architecture |

|
|||||
| Description | VPS10 domain-containing receptor SorCS3 precursor. | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.010148 (rank : 86) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.009628 (rank : 87) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.009601 (rank : 88) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.009013 (rank : 89) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CACB2_MOUSE
|
||||||
| NC score | 0.008625 (rank : 90) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CC27, Q8C5J5, Q9CTQ6 | Gene names | Cacnb2, Cacnlb2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-2 (CAB2) (Calcium channel voltage-dependent subunit beta 2). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.008038 (rank : 91) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
TF3C2_HUMAN
|
||||||
| NC score | 0.007202 (rank : 92) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WUA4, Q16632, Q9BWI7 | Gene names | GTF3C2, KIAA0011 | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor 3C polypeptide 2 (Transcription factor IIIC-subunit beta) (TF3C-beta) (TFIIIC 110 kDa subunit) (TFIIIC110). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.006608 (rank : 93) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.006298 (rank : 94) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
VMDL3_HUMAN
|
||||||
| NC score | 0.005077 (rank : 95) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
DMXL1_MOUSE
|
||||||
| NC score | 0.003971 (rank : 96) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PNC0, Q3TLG8, Q6PHM6, Q8BQF2 | Gene names | Dmxl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
IQEC3_MOUSE
|
||||||
| NC score | 0.003463 (rank : 97) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3TES0, Q3UHP8, Q80TJ8 | Gene names | Iqsec3, Kiaa1110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 3. | |||||
|
APOA_HUMAN
|
||||||
| NC score | 0.003397 (rank : 98) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
PACN2_MOUSE
|
||||||
| NC score | 0.003176 (rank : 99) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVE8 | Gene names | Pacsin2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C and casein kinase substrate in neurons protein 2. | |||||
|
FOXD1_HUMAN
|
||||||
| NC score | 0.002932 (rank : 100) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
KLF14_HUMAN
|
||||||
| NC score | -0.001999 (rank : 101) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD94 | Gene names | KLF14, BTEB5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 14 (Transcription factor BTEB5) (Basic transcription element-binding protein 5) (BTE-binding protein 5). | |||||
|
ZN433_HUMAN
|
||||||
| NC score | -0.002441 (rank : 102) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N7K0 | Gene names | ZNF433 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 433. | |||||
|
ZN148_MOUSE
|
||||||
| NC score | -0.003033 (rank : 103) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 92 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||