Please be patient as the page loads
|
FBX38_HUMAN
|
||||||
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FBX38_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FBX38_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.951686 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 3) | NC score | 0.116483 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 4) | NC score | 0.127339 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 5) | NC score | 0.087729 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.095683 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.126209 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.080248 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 9) | NC score | 0.048533 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.093229 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
WDR60_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.066942 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.089497 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.045556 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.144258 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.091486 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.079410 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.065836 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
LRC29_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.095733 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WV35, Q9UKA0 | Gene names | LRRC29, FBL9, FBXL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 29 (F-box/LRR-repeat protein 9) (F-box and leucine-rich repeat protein 9) (F-box protein FBL9). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.053239 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.026430 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.038730 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDX3X_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.039392 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.039369 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.119512 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.087606 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
ENL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.064253 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
HORN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.072122 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
ABL1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.007745 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.059626 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PL10_MOUSE
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.038401 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
PRP4B_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.013858 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.028418 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.050904 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
DVL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.045560 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |
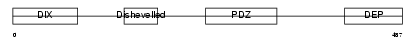
|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
DVL3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 35) | NC score | 0.046172 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
EAF2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.043896 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96CJ1, Q9NZ82 | Gene names | EAF2, TRAITS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 2 (Testosterone-regulated apoptosis inducer and tumor suppressor protein). | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.051475 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.053166 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.028051 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
BASP_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.048945 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
EVL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.039291 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.026783 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.029990 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.040237 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.026959 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.081283 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.014498 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
CCD60_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.046798 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4J0, Q6DIA4, Q6PAQ3 | Gene names | Ccdc60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60. | |||||
|
FBX34_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.060107 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
GCSP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.047711 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23378 | Gene names | GLDC, GCSP | |||
|
Domain Architecture |

|
|||||
| Description | Glycine dehydrogenase [decarboxylating], mitochondrial precursor (EC 1.4.4.2) (Glycine decarboxylase) (Glycine cleavage system P- protein). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.018357 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.078691 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.027662 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
SCG1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.054079 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.029625 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.037980 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
DDIT3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.043506 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
DDX3Y_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.035259 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
EI2BA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.064347 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LC8 | Gene names | Eif2b1 | |||
|
Domain Architecture |
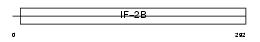
|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.011260 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.075985 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
PININ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 62) | NC score | 0.053217 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
RMP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 63) | NC score | 0.052476 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94763 | Gene names | RMP, C19orf2 | |||
|
Domain Architecture |
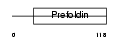
|
|||||
| Description | RNA polymerase II subunit 5-mediating protein (RPB5-mediating protein). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.073503 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
ZN750_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.036800 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
CHFR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.026389 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810L3, Q8BJZ9, Q8BWH4 | Gene names | Chfr | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.091790 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PAPOA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.018972 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.040219 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.024300 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
CF010_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.046071 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
DC1L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.030847 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6G9, Q53HC8, Q53HK7 | Gene names | DYNC1LI1, DNCLI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
DC1L1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.030778 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1Q8 | Gene names | Dync1li1, Dncli1, Dnclic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
HMBX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.027482 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NT76, Q3Y6P1, Q96GS5, Q9H701 | Gene names | HMBOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox-containing protein 1. | |||||
|
HORN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 75) | NC score | 0.059052 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
LDB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 76) | NC score | 0.027819 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70662 | Gene names | Ldb1, Nli | |||
|
Domain Architecture |
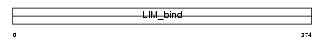
|
|||||
| Description | LIM domain-binding protein 1 (Nuclear LIM interactor). | |||||
|
MYC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.029091 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.018433 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.032032 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.026321 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BOC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.015304 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
BOC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.015341 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 83) | NC score | 0.043006 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
EI2BA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.054510 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14232 | Gene names | EIF2B1, EIF2BA | |||
|
Domain Architecture |
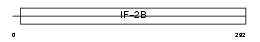
|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
K1849_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.017111 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
MYC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.026571 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.016202 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
NOC2L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.028374 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
PBX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.018404 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40424 | Gene names | PBX1, PRL | |||
|
Domain Architecture |
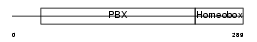
|
|||||
| Description | Pre-B-cell leukemia transcription factor 1 (Homeobox protein PBX1) (Homeobox protein PRL). | |||||
|
PBX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.018404 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41778 | Gene names | Pbx1, Pbx-1 | |||
|
Domain Architecture |
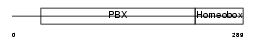
|
|||||
| Description | Pre-B-cell leukemia transcription factor 1 (Homeobox protein PBX1). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.014469 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
ZBT40_HUMAN
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.000488 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
CDSN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.038034 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.034253 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.014521 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
FBX34_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.043701 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWN3, Q2VPB5, Q86TY4 | Gene names | FBXO34, FBX34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.015752 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.037859 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
KLF12_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.001744 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4X4, Q5VZM7, Q9UHZ0 | Gene names | KLF12, AP2REP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 12 (Transcriptional repressor AP-2rep). | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.015660 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
NADAP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.028210 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
NRK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.003896 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
OLIG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.017316 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |
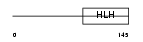
|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.005669 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.025784 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.036866 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CK056_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.019709 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
CTGE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.017789 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.032053 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ING3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.015302 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
P80C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.058551 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.033477 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RHPN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.021342 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUC4, Q8N3T7, Q8N9D6, Q8NE33, Q96RU1 | Gene names | RHPN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhophilin-2 (GTP-Rho-binding protein 2) (76 kDa RhoB effector protein) (p76RBE). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.040996 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.030170 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.015136 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
WTAP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.027241 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ER69 | Gene names | Wtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
3BP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.015489 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.049411 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ANC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.035236 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.027587 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CECR6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.021662 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MX7 | Gene names | Cecr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6 homolog. | |||||
|
CROP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.032882 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.004348 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
I12R2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.012169 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97378 | Gene names | Il12rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-2 chain precursor (IL-12 receptor beta-2) (IL-12R-beta2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.068776 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LDB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.022328 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U70, O96010, Q9UGM4 | Gene names | LDB1, CLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 1 (Carboxyl-terminal LIM domain-binding protein 2) (CLIM-2) (LIM domain-binding factor CLIM2). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.030604 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
OTX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.005144 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
RBP56_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.018344 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.036609 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
ZN406_MOUSE
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.000089 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||
|
E2AK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.003618 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
FXL21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.016567 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKT6 | Gene names | FBXL21, FBL21, FBL3, FBXL3B | |||
|
Domain Architecture |
|
|||||
| Description | F-box/LRR-repeat protein 21 (F-box and leucine-rich repeat protein 21) (F-box/LRR-repeat protein 3B) (F-box and leucine-rich repeat protein 3B). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.016678 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.013738 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
NU153_HUMAN
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.028169 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.017255 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
STIM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.010656 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
TTC14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.016620 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
WTAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.031241 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15007, Q96T28, Q9BYJ7, Q9H4E2 | Gene names | WTAP, KIAA0105 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.051054 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.063358 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.064010 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.076105 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.056281 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.058694 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.080051 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TTC16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.054548 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.056951 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FBX38_MOUSE
|
||||||
| NC score | 0.951686 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.144258 (rank : 3) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.127339 (rank : 4) | θ value | 0.00134147 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.126209 (rank : 5) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.119512 (rank : 6) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.116483 (rank : 7) | θ value | 0.000461057 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
LRC29_HUMAN
|
||||||
| NC score | 0.095733 (rank : 8) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WV35, Q9UKA0 | Gene names | LRRC29, FBL9, FBXL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 29 (F-box/LRR-repeat protein 9) (F-box and leucine-rich repeat protein 9) (F-box protein FBL9). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.095683 (rank : 9) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.093229 (rank : 10) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.091790 (rank : 11) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.091486 (rank : 12) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.089497 (rank : 13) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.087729 (rank : 14) | θ value | 0.00175202 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.087606 (rank : 15) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.081283 (rank : 16) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.080248 (rank : 17) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.080051 (rank : 18) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.079410 (rank : 19) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.078691 (rank : 20) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.076105 (rank : 21) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.075985 (rank : 22) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.073503 (rank : 23) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.072122 (rank : 24) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.068776 (rank : 25) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
WDR60_MOUSE
|
||||||
| NC score | 0.066942 (rank : 26) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C761, Q3UNM4 | Gene names | Wdr60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.065836 (rank : 27) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
EI2BA_MOUSE
|
||||||
| NC score | 0.064347 (rank : 28) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99LC8 | Gene names | Eif2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.064253 (rank : 29) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.064010 (rank : 30) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.063358 (rank : 31) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
FBX34_MOUSE
|
||||||
| NC score | 0.060107 (rank : 32) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.059626 (rank : 33) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.059052 (rank : 34) | θ value | 2.36792 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.058694 (rank : 35) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
P80C_HUMAN
|
||||||
| NC score | 0.058551 (rank : 36) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P38432 | Gene names | COIL, CLN80 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coilin (p80). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.056951 (rank : 37) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.056281 (rank : 38) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
TTC16_HUMAN
|
||||||
| NC score | 0.054548 (rank : 39) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEE8, Q5JU66, Q96M72 | Gene names | TTC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 16 (TPR repeat protein 16). | |||||
|
EI2BA_HUMAN
|
||||||
| NC score | 0.054510 (rank : 40) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14232 | Gene names | EIF2B1, EIF2BA | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.054079 (rank : 41) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.053239 (rank : 42) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.053217 (rank : 43) | θ value | 1.38821 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.053166 (rank : 44) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
RMP_HUMAN
|
||||||
| NC score | 0.052476 (rank : 45) | θ value | 1.38821 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O94763 | Gene names | RMP, C19orf2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit 5-mediating protein (RPB5-mediating protein). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.051475 (rank : 46) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.051054 (rank : 47) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.050904 (rank : 48) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.049411 (rank : 49) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.048945 (rank : 50) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.048533 (rank : 51) | θ value | 0.0148317 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
GCSP_HUMAN
|
||||||
| NC score | 0.047711 (rank : 52) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P23378 | Gene names | GLDC, GCSP | |||
|
Domain Architecture |

|
|||||
| Description | Glycine dehydrogenase [decarboxylating], mitochondrial precursor (EC 1.4.4.2) (Glycine decarboxylase) (Glycine cleavage system P- protein). | |||||
|
CCD60_MOUSE
|
||||||
| NC score | 0.046798 (rank : 53) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C4J0, Q6DIA4, Q6PAQ3 | Gene names | Ccdc60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60. | |||||
|
DVL3_MOUSE
|
||||||
| NC score | 0.046172 (rank : 54) | θ value | 0.47712 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61062 | Gene names | Dvl3 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.046071 (rank : 55) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
DVL3_HUMAN
|
||||||
| NC score | 0.045560 (rank : 56) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92997, O14642, Q13531, Q8N5E9, Q92607 | Gene names | DVL3, KIAA0208 | |||
|
Domain Architecture |

|
|||||
| Description | Segment polarity protein dishevelled homolog DVL-3 (Dishevelled-3) (DSH homolog 3). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.045556 (rank : 57) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
EAF2_HUMAN
|
||||||
| NC score | 0.043896 (rank : 58) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96CJ1, Q9NZ82 | Gene names | EAF2, TRAITS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 2 (Testosterone-regulated apoptosis inducer and tumor suppressor protein). | |||||
|
FBX34_HUMAN
|
||||||
| NC score | 0.043701 (rank : 59) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWN3, Q2VPB5, Q86TY4 | Gene names | FBXO34, FBX34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
DDIT3_MOUSE
|
||||||
| NC score | 0.043506 (rank : 60) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35639, Q91YW9 | Gene names | Ddit3, Chop, Gadd153 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.043006 (rank : 61) | θ value | 3.0926 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.040996 (rank : 62) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.040237 (rank : 63) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.040219 (rank : 64) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
DDX3X_HUMAN
|
||||||
| NC score | 0.039392 (rank : 65) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00571, O15536 | Gene names | DDX3X, DBX, DDX3 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (Helicase-like protein 2) (HLP2) (DEAD box, X isoform). | |||||
|
DDX3X_MOUSE
|
||||||
| NC score | 0.039369 (rank : 66) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62167, O09060, O09143 | Gene names | Ddx3x, D1Pas1-rs2, Ddx3, Dead3, Erh | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3X (EC 3.6.1.-) (DEAD box protein 3, X- chromosomal) (DEAD box RNA helicase DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1Pas1-related sequence 2). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.039291 (rank : 67) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.038730 (rank : 68) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
PL10_MOUSE
|
||||||
| NC score | 0.038401 (rank : 69) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16381 | Gene names | D1Pas1, Pl10 | |||
|
Domain Architecture |

|
|||||
| Description | Putative ATP-dependent RNA helicase Pl10 (EC 3.6.1.-). | |||||
|
CDSN_MOUSE
|
||||||
| NC score | 0.038034 (rank : 70) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TPC1 | Gene names | Cdsn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Corneodesmosin precursor. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.037980 (rank : 71) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.037859 (rank : 72) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.036866 (rank : 73) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ZN750_HUMAN
|
||||||
| NC score | 0.036800 (rank : 74) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q32MQ0, Q9H899 | Gene names | ZNF750 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF750. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.036609 (rank : 75) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
DDX3Y_HUMAN
|
||||||
| NC score | 0.035259 (rank : 76) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15523, Q8IYV7 | Gene names | DDX3Y, DBY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX3Y (EC 3.6.1.-) (DEAD box protein 3, Y- chromosomal). | |||||
|
ANC1_MOUSE
|
||||||
| NC score | 0.035236 (rank : 77) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.034253 (rank : 78) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.033477 (rank : 79) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
CROP_MOUSE
|
||||||
| NC score | 0.032882 (rank : 80) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5SUF2, Q3U9D5, Q8BUJ5, Q921Z3, Q9CRS7, Q9CTY4 | Gene names | Crop | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cisplatin resistance-associated overexpressed protein. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.032053 (rank : 81) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.032032 (rank : 82) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
WTAP_HUMAN
|
||||||
| NC score | 0.031241 (rank : 83) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15007, Q96T28, Q9BYJ7, Q9H4E2 | Gene names | WTAP, KIAA0105 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
DC1L1_HUMAN
|
||||||
| NC score | 0.030847 (rank : 84) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6G9, Q53HC8, Q53HK7 | Gene names | DYNC1LI1, DNCLI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
DC1L1_MOUSE
|
||||||
| NC score | 0.030778 (rank : 85) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R1Q8 | Gene names | Dync1li1, Dncli1, Dnclic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic dynein 1 light intermediate chain 1 (Dynein light intermediate chain 1, cytosolic) (Dynein light chain A) (DLC-A). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.030604 (rank : 86) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.030170 (rank : 87) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.029990 (rank : 88) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.029625 (rank : 89) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
MYC_HUMAN
|
||||||
| NC score | 0.029091 (rank : 90) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01106, P01107, Q14026 | Gene names | MYC | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.028418 (rank : 91) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
NOC2L_HUMAN
|
||||||
| NC score | 0.028374 (rank : 92) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3T9, Q5SVA3, Q9BTN6 | Gene names | NOC2L | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar complex protein 2 homolog (Protein NOC2 homolog) (NOC2- like). | |||||
|
NADAP_HUMAN
|
||||||
| NC score | 0.028210 (rank : 93) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BWU0, Q4KMT1, Q4KMX0, Q7Z5Q9, Q9NVN2 | Gene names | SLC4A1AP, HLC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kanadaptin (Kidney anion exchanger adapter protein) (Solute carrier family 4 anion exchanger member 1 adapter protein) (Lung cancer oncogene 3 protein). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.028169 (rank : 94) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.028051 (rank : 95) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
LDB1_MOUSE
|
||||||
| NC score | 0.027819 (rank : 96) | θ value | 2.36792 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70662 | Gene names | Ldb1, Nli | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-binding protein 1 (Nuclear LIM interactor). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.027662 (rank : 97) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.027587 (rank : 98) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
HMBX1_HUMAN
|
||||||
| NC score | 0.027482 (rank : 99) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NT76, Q3Y6P1, Q96GS5, Q9H701 | Gene names | HMBOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox-containing protein 1. | |||||
|
WTAP_MOUSE
|
||||||
| NC score | 0.027241 (rank : 100) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ER69 | Gene names | Wtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.026959 (rank : 101) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.026783 (rank : 102) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.026571 (rank : 103) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.026430 (rank : 104) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CHFR_MOUSE
|
||||||
| NC score | 0.026389 (rank : 105) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q810L3, Q8BJZ9, Q8BWH4 | Gene names | Chfr | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase CHFR (EC 6.3.2.-) (Checkpoint with forkhead and RING finger domains protein). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.026321 (rank : 106) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.025784 (rank : 107) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.024300 (rank : 108) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
LDB1_HUMAN
|
||||||
| NC score | 0.022328 (rank : 109) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86U70, O96010, Q9UGM4 | Gene names | LDB1, CLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 1 (Carboxyl-terminal LIM domain-binding protein 2) (CLIM-2) (LIM domain-binding factor CLIM2). | |||||
|
CECR6_MOUSE
|
||||||
| NC score | 0.021662 (rank : 110) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MX7 | Gene names | Cecr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cat eye syndrome critical region protein 6 homolog. | |||||
|
RHPN2_HUMAN
|
||||||
| NC score | 0.021342 (rank : 111) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUC4, Q8N3T7, Q8N9D6, Q8NE33, Q96RU1 | Gene names | RHPN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhophilin-2 (GTP-Rho-binding protein 2) (76 kDa RhoB effector protein) (p76RBE). | |||||
|
CK056_HUMAN
|
||||||
| NC score | 0.019709 (rank : 112) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N612, Q9C0A4, Q9H0N3, Q9H624 | Gene names | C11orf56, KIAA1759 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf56. | |||||
|
PAPOA_HUMAN
|
||||||
| NC score | 0.018972 (rank : 113) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51003, Q86SX4, Q86TV0, Q8IYF5, Q9BVU2 | Gene names | PAPOLA, PAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(A) polymerase alpha (EC 2.7.7.19) (PAP) (Polynucleotide adenylyltransferase alpha). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.018433 (rank : 114) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
PBX1_HUMAN
|
||||||
| NC score | 0.018404 (rank : 115) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40424 | Gene names | PBX1, PRL | |||
|
Domain Architecture |

|
|||||
| Description | Pre-B-cell leukemia transcription factor 1 (Homeobox protein PBX1) (Homeobox protein PRL). | |||||
|
PBX1_MOUSE
|
||||||
| NC score | 0.018404 (rank : 116) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P41778 | Gene names | Pbx1, Pbx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Pre-B-cell leukemia transcription factor 1 (Homeobox protein PBX1). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.018357 (rank : 117) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
RBP56_HUMAN
|
||||||
| NC score | 0.018344 (rank : 118) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92804, Q92751 | Gene names | TAF15, RBP56, TAF2N | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated factor 2N (RNA-binding protein 56) (TAFII68) (TAF(II)68). | |||||
|
CTGE2_HUMAN
|
||||||
| NC score | 0.017789 (rank : 119) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
OLIG1_HUMAN
|
||||||
| NC score | 0.017316 (rank : 120) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAK6, Q7RTS0 | Gene names | OLIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 1 (Oligo1). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.017255 (rank : 121) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.017111 (rank : 122) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.016678 (rank : 123) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
TTC14_HUMAN
|
||||||
| NC score | 0.016620 (rank : 124) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96N46, Q6UWJ7, Q8TF22 | Gene names | TTC14, KIAA1980 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 14 (TPR repeat protein 14). | |||||
|
FXL21_HUMAN
|
||||||
| NC score | 0.016567 (rank : 125) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKT6 | Gene names | FBXL21, FBL21, FBL3, FBXL3B | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 21 (F-box and leucine-rich repeat protein 21) (F-box/LRR-repeat protein 3B) (F-box and leucine-rich repeat protein 3B). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.016202 (rank : 126) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.015752 (rank : 127) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.015660 (rank : 128) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
3BP5_MOUSE
|
||||||
| NC score | 0.015489 (rank : 129) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
BOC_MOUSE
|
||||||
| NC score | 0.015341 (rank : 130) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AZB0, Q6KAM5, Q6P5H3, Q7TMJ3, Q8CE73, Q8CE91, Q8R377, Q923W7 | Gene names | Boc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
BOC_HUMAN
|
||||||
| NC score | 0.015304 (rank : 131) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
ING3_MOUSE
|
||||||
| NC score | 0.015302 (rank : 132) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.015136 (rank : 133) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.014521 (rank : 134) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.014498 (rank : 135) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.014469 (rank : 136) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
PRP4B_MOUSE
|
||||||
| NC score | 0.013858 (rank : 137) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61136, O88378, Q8BND8, Q8R4Y5, Q9CTL9, Q9CTT0 | Gene names | Prpf4b, Cbp143, Prp4h, Prp4k, Prp4m, Prpk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PRP4 homolog (EC 2.7.11.1) (PRP4 pre- mRNA-processing factor 4 homolog) (Pre-mRNA protein kinase). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.013738 (rank : 138) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
I12R2_MOUSE
|
||||||
| NC score | 0.012169 (rank : 139) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97378 | Gene names | Il12rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-12 receptor beta-2 chain precursor (IL-12 receptor beta-2) (IL-12R-beta2). | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.011260 (rank : 140) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
STIM1_MOUSE
|
||||||
| NC score | 0.010656 (rank : 141) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70302 | Gene names | Stim1, Sim | |||
|
Domain Architecture |

|
|||||
| Description | Stromal interaction molecule 1 precursor. | |||||
|
ABL1_MOUSE
|
||||||
| NC score | 0.007745 (rank : 142) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P00520, P97896, Q61252, Q61253, Q61254, Q61255, Q61256, Q61257, Q61258, Q61259, Q61260, Q61261, Q6PCM5, Q8C1X4 | Gene names | Abl1, Abl | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.005669 (rank : 143) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
OTX1_MOUSE
|
||||||
| NC score | 0.005144 (rank : 144) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80205 | Gene names | Otx1, Otx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein OTX1. | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.004348 (rank : 145) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
NRK_MOUSE
|
||||||
| NC score | 0.003896 (rank : 146) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9R0G8, Q6NV55, Q8C9S9, Q9R0S4 | Gene names | Nrk, Nesk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1) (Nck-interacting kinase-like embryo specific kinase) (NIK-like embryo-specific kinase) (NESK). | |||||
|
E2AK4_HUMAN
|
||||||
| NC score | 0.003618 (rank : 147) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1012 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2K8, Q69YL7, Q6DC97, Q96GN6, Q9H5K1, Q9NSQ3, Q9NSZ5, Q9UJ56 | Gene names | EIF2AK4, GCN2, KIAA1338 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 4 (EC 2.7.11.1) (GCN2-like protein). | |||||
|
KLF12_HUMAN
|
||||||
| NC score | 0.001744 (rank : 148) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y4X4, Q5VZM7, Q9UHZ0 | Gene names | KLF12, AP2REP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 12 (Transcriptional repressor AP-2rep). | |||||
|
ZBT40_HUMAN
|
||||||
| NC score | 0.000488 (rank : 149) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NUA8, O75066, Q5TFU5, Q8N1R1 | Gene names | ZBTB40, KIAA0478 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 40. | |||||
|
ZN406_MOUSE
|
||||||
| NC score | 0.000089 (rank : 150) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TS63 | Gene names | Znf406, Zfp406 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 406. | |||||