Please be patient as the page loads
|
FBX34_MOUSE
|
||||||
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FBX34_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975093 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NWN3, Q2VPB5, Q86TY4 | Gene names | FBXO34, FBX34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
FBX34_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
FBX46_HUMAN
|
||||||
| θ value | 3.23591e-74 (rank : 3) | NC score | 0.867684 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PJ61 | Gene names | FBXO46, FBX46, FBXO34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 8.81223e-72 (rank : 4) | NC score | 0.849997 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 5) | NC score | 0.050846 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.044780 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
BRWD1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.025988 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.064709 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.037477 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.030444 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.047123 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.060635 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
DACH2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.036810 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NX9, Q8NAY3, Q8ND17, Q96N55 | Gene names | DACH2 | |||
|
Domain Architecture |
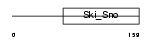
|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
FXR2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.027411 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.015337 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.010094 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.034665 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
FXL21_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.039202 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKT6 | Gene names | FBXL21, FBL21, FBL3, FBXL3B | |||
|
Domain Architecture |
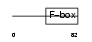
|
|||||
| Description | F-box/LRR-repeat protein 21 (F-box and leucine-rich repeat protein 21) (F-box/LRR-repeat protein 3B) (F-box and leucine-rich repeat protein 3B). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.042427 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
FBX38_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.060107 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FBX38_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.059924 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.019644 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
NEK4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.007397 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51957 | Gene names | NEK4, STK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2) (Serine/threonine-protein kinase NRK2). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.028854 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.038373 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CMTA2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.039327 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.009458 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.034919 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
ACINU_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.033537 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ACRBP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.031535 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3V140, Q62253, Q62254, Q8C621, Q91VQ1 | Gene names | Acrbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acrosin-binding protein precursor (Proacrosin-binding protein sp32). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.021839 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.030574 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
FXL21_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.034431 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BFZ4, Q8BIJ8, Q8BIW5 | Gene names | Fbxl21, Fbl21, Fbxl3b | |||
|
Domain Architecture |
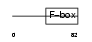
|
|||||
| Description | F-box/LRR-repeat protein 21 (F-box and leucine-rich repeat protein 21) (F-box/LRR-repeat protein 3B) (F-box and leucine-rich repeat protein 3B). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.012117 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SOX9_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.024191 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
SP7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.003252 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDD2 | Gene names | SP7, OSX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp7 (Zinc finger protein osterix). | |||||
|
TEC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.007894 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P24604, Q9R1M9, Q9WVN0, Q9WVN1, Q9WVN2, Q9WVN3 | Gene names | Tec | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
TRI47_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.014774 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
WEE1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.009693 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
ABCA9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.008656 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUA7, Q6P655, Q8N2S4, Q8WWZ5, Q96MD8 | Gene names | ABCA9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 9. | |||||
|
BSCL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.037257 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
NOL4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.022991 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60954 | Gene names | Nol4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4. | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.032703 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
ASPX_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.030345 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.026187 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.013077 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.017116 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.015094 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.020000 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.006209 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
WWP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.026670 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.028610 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CD68_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.024205 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P34810, Q96BI7 | Gene names | CD68 | |||
|
Domain Architecture |
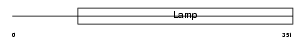
|
|||||
| Description | Macrosialin precursor (GP110) (CD68 antigen). | |||||
|
IF39_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.019601 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
IF4G3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.015645 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MCE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.026911 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55236 | Gene names | Rngtt, Cap1a | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (MCE1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.018479 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.007314 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RAE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.014594 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.013262 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
REXO1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.018913 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
STK10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.005966 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SYT10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.010975 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
TEC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.005954 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42680 | Gene names | TEC, PSCTK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
ZN509_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.000244 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.007106 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
ARHG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.011400 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.019335 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CTND2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.013320 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
DRAP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.022513 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.017399 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.015905 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.009461 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.026139 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CEND3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.006229 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.011036 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.013898 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
LIN9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.015616 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TKA1, Q5U5L8, Q5U7E1, Q6PI55, Q6ZTV4, Q7Z3J1 | Gene names | LIN9, BARA, TGS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-9 homolog (huLin-9) (hLin-9) (Beta-subunit associated regulator of apoptosis) (Type I interferon receptor beta chain-associated protein) (TUDOR gene similar protein) (pRB-associated protein). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.007399 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.009788 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
MCE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.023946 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60942, O43483, O60257, O60351 | Gene names | RNGTT, CAP1A | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (HCAP1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.011323 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
RAB3I_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.014939 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
SHC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.012375 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92529, Q8TAP2, Q9UCX5 | Gene names | SHC3, NSHC, SHCC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc) (Protein Rai). | |||||
|
SHC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.012731 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61120 | Gene names | Shc3, Nshc, ShcC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc). | |||||
|
SYT6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.009878 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |
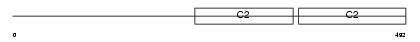
|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
TACC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.011414 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.006547 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.005823 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CRTC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.013534 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CTND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.012982 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.021277 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.013915 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.018079 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
M4K4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.004304 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.007339 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.004341 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
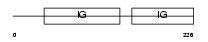
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.012050 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.010200 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
ZN691_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | -0.000688 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VV52, O95878, Q9NWE8 | Gene names | ZNF691 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 691. | |||||
|
FBX34_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q80XI1, Q9D290 | Gene names | Fbxo34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
FBX34_HUMAN
|
||||||
| NC score | 0.975093 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NWN3, Q2VPB5, Q86TY4 | Gene names | FBXO34, FBX34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 34. | |||||
|
FBX46_HUMAN
|
||||||
| NC score | 0.867684 (rank : 3) | θ value | 3.23591e-74 (rank : 3) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PJ61 | Gene names | FBXO46, FBX46, FBXO34L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.849997 (rank : 4) | θ value | 8.81223e-72 (rank : 4) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.064709 (rank : 5) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.060635 (rank : 6) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.060107 (rank : 7) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FBX38_MOUSE
|
||||||
| NC score | 0.059924 (rank : 8) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.050846 (rank : 9) | θ value | 0.0193708 (rank : 5) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.047123 (rank : 10) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.044780 (rank : 11) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.042427 (rank : 12) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CMTA2_MOUSE
|
||||||
| NC score | 0.039327 (rank : 13) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80Y50, Q3TFK0, Q5SX68, Q80TP1, Q8R0D9, Q8R2N5 | Gene names | Camta2, Kiaa0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
FXL21_HUMAN
|
||||||
| NC score | 0.039202 (rank : 14) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKT6 | Gene names | FBXL21, FBL21, FBL3, FBXL3B | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 21 (F-box and leucine-rich repeat protein 21) (F-box/LRR-repeat protein 3B) (F-box and leucine-rich repeat protein 3B). | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.038373 (rank : 15) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.037477 (rank : 16) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
BSCL2_HUMAN
|
||||||
| NC score | 0.037257 (rank : 17) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96G97, Q96SV1, Q9BSQ0 | Gene names | BSCL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Seipin (Bernardinelli-Seip congenital lipodystrophy type 2 protein). | |||||
|
DACH2_HUMAN
|
||||||
| NC score | 0.036810 (rank : 18) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NX9, Q8NAY3, Q8ND17, Q96N55 | Gene names | DACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 2 (Dach2). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.034919 (rank : 19) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.034665 (rank : 20) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
FXL21_MOUSE
|
||||||
| NC score | 0.034431 (rank : 21) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BFZ4, Q8BIJ8, Q8BIW5 | Gene names | Fbxl21, Fbl21, Fbxl3b | |||
|
Domain Architecture |

|
|||||
| Description | F-box/LRR-repeat protein 21 (F-box and leucine-rich repeat protein 21) (F-box/LRR-repeat protein 3B) (F-box and leucine-rich repeat protein 3B). | |||||
|
ACINU_MOUSE
|
||||||
| NC score | 0.033537 (rank : 22) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JIX8, Q9CSN7, Q9CSR9, Q9CSX7, Q9R046, Q9R047 | Gene names | Acin1, Acinus | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.032703 (rank : 23) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
ACRBP_MOUSE
|
||||||
| NC score | 0.031535 (rank : 24) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3V140, Q62253, Q62254, Q8C621, Q91VQ1 | Gene names | Acrbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acrosin-binding protein precursor (Proacrosin-binding protein sp32). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.030574 (rank : 25) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.030444 (rank : 26) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
ASPX_MOUSE
|
||||||
| NC score | 0.030345 (rank : 27) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50289, Q9DAM6 | Gene names | Acrv1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1) (MSA- 63). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.028854 (rank : 28) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.028610 (rank : 29) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
FXR2_HUMAN
|
||||||
| NC score | 0.027411 (rank : 30) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51116, Q8WUM2 | Gene names | FXR2 | |||
|
Domain Architecture |

|
|||||
| Description | Fragile X mental retardation syndrome-related protein 2. | |||||
|
MCE1_MOUSE
|
||||||
| NC score | 0.026911 (rank : 31) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55236 | Gene names | Rngtt, Cap1a | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (MCE1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
WWP2_HUMAN
|
||||||
| NC score | 0.026670 (rank : 32) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00308, Q96CZ2, Q9BWN6 | Gene names | WWP2 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-like E3 ubiquitin-protein ligase WWP2 (EC 6.3.2.-) (WW domain- containing protein 2) (Atrophin-1-interacting protein 2) (AIP2). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.026187 (rank : 33) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.026139 (rank : 34) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
BRWD1_MOUSE
|
||||||
| NC score | 0.025988 (rank : 35) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q921C3, Q921C2 | Gene names | Brwd1, Wdr9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and WD repeat domain-containing protein 1 (WD repeat protein 9). | |||||
|
CD68_HUMAN
|
||||||
| NC score | 0.024205 (rank : 36) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P34810, Q96BI7 | Gene names | CD68 | |||
|
Domain Architecture |

|
|||||
| Description | Macrosialin precursor (GP110) (CD68 antigen). | |||||
|
SOX9_MOUSE
|
||||||
| NC score | 0.024191 (rank : 37) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04887, Q8C7L2, Q91ZK2, Q99KQ0 | Gene names | Sox9, Sox-9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
MCE1_HUMAN
|
||||||
| NC score | 0.023946 (rank : 38) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60942, O43483, O60257, O60351 | Gene names | RNGTT, CAP1A | |||
|
Domain Architecture |

|
|||||
| Description | mRNA capping enzyme (HCE) (HCAP1) [Includes: Polynucleotide 5'- triphosphatase (EC 3.1.3.33) (mRNA 5'-triphosphatase) (TPase); mRNA guanylyltransferase (EC 2.7.7.50) (GTP--RNA guanylyltransferase) (GTase)]. | |||||
|
NOL4_MOUSE
|
||||||
| NC score | 0.022991 (rank : 39) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P60954 | Gene names | Nol4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 4. | |||||
|
DRAP1_MOUSE
|
||||||
| NC score | 0.022513 (rank : 40) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.021839 (rank : 41) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.021277 (rank : 42) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.020000 (rank : 43) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.019644 (rank : 44) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
IF39_HUMAN
|
||||||
| NC score | 0.019601 (rank : 45) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P55884, Q9UMF9 | Gene names | EIF3S9 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 9 (eIF-3 eta) (eIF3 p116) (eIF3 p110) (eIF3b) (Prt1 homolog) (hPrt1). | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.019335 (rank : 46) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
REXO1_HUMAN
|
||||||
| NC score | 0.018913 (rank : 47) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1G1, Q9ULT2 | Gene names | REXO1, ELOABP1, KIAA1138, TCEB3BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA exonuclease 1 homolog (EC 3.1.-.-) (Elongin A-binding protein 1) (EloA-BP1) (Transcription elongation factor B polypeptide 3-binding protein 1). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.018479 (rank : 48) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.018079 (rank : 49) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.017399 (rank : 50) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.017116 (rank : 51) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.015905 (rank : 52) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
IF4G3_MOUSE
|
||||||
| NC score | 0.015645 (rank : 53) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80XI3, Q80Y69, Q8BJ68, Q8BQF3, Q8C6Q3, Q8CIH0 | Gene names | Eif4g3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
LIN9_HUMAN
|
||||||
| NC score | 0.015616 (rank : 54) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5TKA1, Q5U5L8, Q5U7E1, Q6PI55, Q6ZTV4, Q7Z3J1 | Gene names | LIN9, BARA, TGS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-9 homolog (huLin-9) (hLin-9) (Beta-subunit associated regulator of apoptosis) (Type I interferon receptor beta chain-associated protein) (TUDOR gene similar protein) (pRB-associated protein). | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.015337 (rank : 55) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.015094 (rank : 56) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RAB3I_HUMAN
|
||||||
| NC score | 0.014939 (rank : 57) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96QF0, Q6PCE4, Q96A24, Q96QE6, Q96QE7, Q96QE8, Q96QE9, Q96QF1, Q9H673 | Gene names | RAB3IP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAB3A-interacting protein (Rabin-3) (SSX2-interacting protein). | |||||
|
TRI47_MOUSE
|
||||||
| NC score | 0.014774 (rank : 58) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
RAE1_MOUSE
|
||||||
| NC score | 0.014594 (rank : 59) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QXG2, Q80UV8 | Gene names | Chm, Rep1 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 1 (Rab escort protein 1) (REP-1) (Choroideraemia protein homolog). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.013915 (rank : 60) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.013898 (rank : 61) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
CRTC1_MOUSE
|
||||||
| NC score | 0.013534 (rank : 62) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q68ED7, Q6ZQ85 | Gene names | Crtc1, Kiaa0616, Mect1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CREB-regulated transcription coactivator 1 (Mucoepidermoid carcinoma translocated protein 1 homolog). | |||||
|
CTND2_MOUSE
|
||||||
| NC score | 0.013320 (rank : 63) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O35927 | Gene names | Ctnnd2, Catnd2, Nprap | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin). | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.013262 (rank : 64) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.013077 (rank : 65) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CTND2_HUMAN
|
||||||
| NC score | 0.012982 (rank : 66) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UQB3, O00379, O15390, O43206, O43840, Q13589, Q9UM66, Q9UPM3 | Gene names | CTNND2, NPRAP | |||
|
Domain Architecture |

|
|||||
| Description | Catenin delta-2 (Delta-catenin) (Neural plakophilin-related ARM-repeat protein) (NPRAP) (Neurojungin) (GT24). | |||||
|
SHC3_MOUSE
|
||||||
| NC score | 0.012731 (rank : 67) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61120 | Gene names | Shc3, Nshc, ShcC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc). | |||||
|
SHC3_HUMAN
|
||||||
| NC score | 0.012375 (rank : 68) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92529, Q8TAP2, Q9UCX5 | Gene names | SHC3, NSHC, SHCC | |||
|
Domain Architecture |

|
|||||
| Description | SHC-transforming protein 3 (SH2 domain protein C3) (Src homology 2 domain-containing-transforming protein C3) (Neuronal Shc) (N-Shc) (Protein Rai). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.012117 (rank : 69) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.012050 (rank : 70) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TACC1_HUMAN
|
||||||
| NC score | 0.011414 (rank : 71) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75410, Q6Y687, Q86YG7, Q8IUJ2, Q8IUJ3, Q8IUJ4, Q8IZG2, Q8NEY7, Q9UPP9 | Gene names | TACC1, KIAA1103 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 1 (Taxin 1) (Gastric cancer antigen Ga55). | |||||
|
ARHG1_MOUSE
|
||||||
| NC score | 0.011400 (rank : 72) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61210, O89074, Q80YE8, Q80YE9, Q91VL3 | Gene names | Arhgef1, Lbcl2, Lsc | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 1 (Lymphoid blast crisis-like 2) (Lbc's second cousin). | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.011323 (rank : 73) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.011036 (rank : 74) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
SYT10_HUMAN
|
||||||
| NC score | 0.010975 (rank : 75) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.010200 (rank : 76) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.010094 (rank : 77) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
SYT6_MOUSE
|
||||||
| NC score | 0.009878 (rank : 78) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.009788 (rank : 79) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
WEE1_MOUSE
|
||||||
| NC score | 0.009693 (rank : 80) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P47810, Q9EPL7 | Gene names | Wee1 | |||
|
Domain Architecture |

|
|||||
| Description | Wee1-like protein kinase (EC 2.7.10.2) (Wee1A kinase). | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.009461 (rank : 81) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.009458 (rank : 82) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ABCA9_HUMAN
|
||||||
| NC score | 0.008656 (rank : 83) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUA7, Q6P655, Q8N2S4, Q8WWZ5, Q96MD8 | Gene names | ABCA9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 9. | |||||
|
TEC_MOUSE
|
||||||
| NC score | 0.007894 (rank : 84) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P24604, Q9R1M9, Q9WVN0, Q9WVN1, Q9WVN2, Q9WVN3 | Gene names | Tec | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.007399 (rank : 85) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
NEK4_HUMAN
|
||||||
| NC score | 0.007397 (rank : 86) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51957 | Gene names | NEK4, STK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2) (Serine/threonine-protein kinase NRK2). | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.007339 (rank : 87) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.007314 (rank : 88) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.007106 (rank : 89) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.006547 (rank : 90) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
CEND3_MOUSE
|
||||||
| NC score | 0.006229 (rank : 91) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R5G7, Q5DTN4, Q6NVF1, Q8R5G6 | Gene names | Centd3, Arap3, Drag1, Kiaa4097 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-delta 3 (Cnt-d3) (Arf-GAP, Rho-GAP, ankyrin repeat and pleckstrin homology domain-containing protein 3) (Dual specificity Rho- and Arf-GTPase-activating protein 1). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.006209 (rank : 92) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.005966 (rank : 93) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TEC_HUMAN
|
||||||
| NC score | 0.005954 (rank : 94) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P42680 | Gene names | TEC, PSCTK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.005823 (rank : 95) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.004341 (rank : 96) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
M4K4_HUMAN
|
||||||
| NC score | 0.004304 (rank : 97) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 1589 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95819, O75172, Q9NST7 | Gene names | MAP4K4, HGK, KIAA0687, NIK | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
SP7_HUMAN
|
||||||
| NC score | 0.003252 (rank : 98) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TDD2 | Gene names | SP7, OSX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp7 (Zinc finger protein osterix). | |||||
|
ZN509_HUMAN
|
||||||
| NC score | 0.000244 (rank : 99) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZSB9, Q59FJ4, Q5EBN0, Q8TB80 | Gene names | ZNF509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ZN691_HUMAN
|
||||||
| NC score | -0.000688 (rank : 100) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 100 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5VV52, O95878, Q9NWE8 | Gene names | ZNF691 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 691. | |||||