Please be patient as the page loads
|
OXLA_MOUSE
|
||||||
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OXLA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.980060 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
OXLA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
AOFB_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 3) | NC score | 0.502707 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 4) | NC score | 0.517341 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 5) | NC score | 0.499876 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOF1_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 6) | NC score | 0.428039 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
LSD1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 7) | NC score | 0.379553 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 8) | NC score | 0.397070 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
AOF1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 9) | NC score | 0.426153 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOFA_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.463964 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
PAOX_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.318797 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
ERG1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.153303 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |
|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
PAOX_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.289788 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
FMO2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.117796 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K2I3, Q9QZF7 | Gene names | Fmo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2). | |||||
|
FMO3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.110896 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31513, Q14854, Q8N5N5 | Gene names | FMO3 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3) (FMO form 2) (FMO II). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.034202 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
FMO2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.109352 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99518, Q5EBX4, Q9BRX1 | Gene names | FMO2 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2) (FMO 1B1). | |||||
|
ERG1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.135881 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
PPOX_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.181077 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
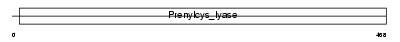
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
P3H3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.035114 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
SMOX_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.268956 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.267192 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
CJ033_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.128155 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
FBX38_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.040237 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FMO3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.114771 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97501 | Gene names | Fmo3 | |||
|
Domain Architecture |
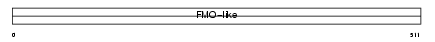
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3). | |||||
|
DLDH_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.068093 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.067983 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
CB025_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.044673 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LS1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf25 homolog, mitochondrial precursor. | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.030881 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
FMO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.102998 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01740, Q5QPT2, Q9UJC2 | Gene names | FMO1 | |||
|
Domain Architecture |
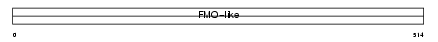
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Fetal hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
FMO1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.095893 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50285 | Gene names | Fmo1, Fmo-1 | |||
|
Domain Architecture |
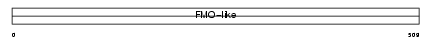
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
FMO4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.094711 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31512 | Gene names | FMO4, FMO2 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
PPOX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.119316 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
UN45A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.027448 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.026776 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
CHDH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.033027 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJ64, Q8CHT7 | Gene names | Chdh | |||
|
Domain Architecture |

|
|||||
| Description | Choline dehydrogenase, mitochondrial precursor (EC 1.1.99.1) (CHD) (CDH). | |||||
|
CJ033_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.106997 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.011537 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
CB025_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.038686 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3L0, O95891 | Gene names | C2orf25, CL25022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf25, mitochondrial precursor. | |||||
|
FLVC2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.018655 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X85 | Gene names | Flvcr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
FMO4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.081412 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHG0 | Gene names | Fmo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.010814 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
GSHR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.045964 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
P3H3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.025821 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
PCYOX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.069427 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
PHLA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.018126 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
TCAL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.023771 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
FMO5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.078919 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49326 | Gene names | FMO5 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 5) (FMO 5) (Dimethylaniline oxidase 5). | |||||
|
FMO5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.074270 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97872 | Gene names | Fmo5 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 5) (FMO 5) (Dimethylaniline oxidase 5). | |||||
|
FMO6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.082677 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60774 | Gene names | FMO6 | |||
|
Domain Architecture |
|
|||||
| Description | Putative dimethylaniline monooxygenase [N-oxide-forming] 6 (EC 1.14.13.8) (Flavin-containing monooxygenase 6) (FMO 6) (Dimethylaniline oxidase 6). | |||||
|
OXLA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O09046, Q3T9N9, Q3U7S6, Q3V0K2, Q6Y632, Q6YBV6, Q6YDI8, Q8R2G8, Q9CXK7 | Gene names | Il4i1, Fig1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (mFIG1). | |||||
|
OXLA_HUMAN
|
||||||
| NC score | 0.980060 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96RQ9, Q4GZN1, Q4GZN2, Q6P2Q3, Q8TEM5, Q96RQ8 | Gene names | IL4I1, FIG1 | |||
|
Domain Architecture |

|
|||||
| Description | L-amino-acid oxidase precursor (EC 1.4.3.2) (LAAO) (Interleukin-4- induced protein 1) (IL4-induced protein 1) (Protein Fig-1) (hFIG1). | |||||
|
AOFB_HUMAN
|
||||||
| NC score | 0.517341 (rank : 3) | θ value | 1.33837e-11 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27338, Q7Z6S2 | Gene names | MAOB | |||
|
Domain Architecture |
|
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFB_MOUSE
|
||||||
| NC score | 0.502707 (rank : 4) | θ value | 4.59992e-12 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BW75, Q8C0B2 | Gene names | Maob | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] B (EC 1.4.3.4) (Monoamine oxidase type B) (MAO-B). | |||||
|
AOFA_MOUSE
|
||||||
| NC score | 0.499876 (rank : 5) | θ value | 4.30538e-10 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q64133, Q8K0Z8 | Gene names | Maoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOFA_HUMAN
|
||||||
| NC score | 0.463964 (rank : 6) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21397, Q16426 | Gene names | MAOA | |||
|
Domain Architecture |

|
|||||
| Description | Amine oxidase [flavin-containing] A (EC 1.4.3.4) (Monoamine oxidase type A) (MAO-A). | |||||
|
AOF1_MOUSE
|
||||||
| NC score | 0.428039 (rank : 7) | θ value | 3.64472e-09 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CIG3, Q8C5C4, Q8CEC1 | Gene names | Aof1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
AOF1_HUMAN
|
||||||
| NC score | 0.426153 (rank : 8) | θ value | 4.45536e-07 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8NB78, Q5TGV3, Q6AI15, Q6ZUU4, Q8N258, Q96EL7 | Gene names | AOF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Flavin-containing amine oxidase domain-containing protein 1 (EC 1.-.-.-). | |||||
|
LSD1_MOUSE
|
||||||
| NC score | 0.397070 (rank : 9) | θ value | 1.53129e-07 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6ZQ88, Q6PB53, Q8VEA1 | Gene names | Aof2, Kiaa0601, Lsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
LSD1_HUMAN
|
||||||
| NC score | 0.379553 (rank : 10) | θ value | 1.53129e-07 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60341, Q5TH94, Q5TH95, Q86VT7, Q8IXK4, Q8NDP6, Q8TAZ3, Q96AW4 | Gene names | AOF2, KIAA0601, LSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysine-specific histone demethylase 1 (EC 1.-.-.-) (Flavin-containing amine oxidase domain-containing protein 2) (BRAF35-HDAC complex protein BHC110). | |||||
|
PAOX_MOUSE
|
||||||
| NC score | 0.318797 (rank : 11) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0L6, Q8K254 | Gene names | Paox, Pao | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
PAOX_HUMAN
|
||||||
| NC score | 0.289788 (rank : 12) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6QHF9, Q6QHF5, Q6QHF6, Q6QHF7, Q6QHF8, Q6QHG0, Q6QHG1, Q6QHG2, Q6QHG3, Q6QHG4, Q6QHG5, Q6QHG6, Q86WP9, Q8N555, Q8NCX3 | Gene names | PAOX, PAO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal N1-acetyl-spermine/spermidine oxidase (EC 1.5.3.11) (Polyamine oxidase). | |||||
|
SMOX_HUMAN
|
||||||
| NC score | 0.268956 (rank : 13) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NWM0, Q5TE26, Q5TE27, Q6UY28, Q8IX00, Q96LC3, Q96LC4, Q96QT3, Q9BW38, Q9H6H1, Q9NP51, Q9NPY1, Q9NPY2 | Gene names | SMOX, C20orf16, SMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
SMOX_MOUSE
|
||||||
| NC score | 0.267192 (rank : 14) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99K82, Q70LA3, Q70LA4, Q70LA5, Q70LA7, Q70LA8, Q70LA9, Q70LB0, Q8CJ56, Q8CJ57 | Gene names | Smox, Smo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermine oxidase (EC 1.5.3.-) (Polyamine oxidase 1) (PAO-1) (PAOh1). | |||||
|
PPOX_HUMAN
|
||||||
| NC score | 0.181077 (rank : 15) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P50336 | Gene names | PPOX | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
ERG1_MOUSE
|
||||||
| NC score | 0.153303 (rank : 16) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P52019 | Gene names | Sqle, Erg1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
ERG1_HUMAN
|
||||||
| NC score | 0.135881 (rank : 17) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14534, Q9UEK6 | Gene names | SQLE, ERG1 | |||
|
Domain Architecture |

|
|||||
| Description | Squalene monooxygenase (EC 1.14.99.7) (Squalene epoxidase) (SE). | |||||
|
CJ033_MOUSE
|
||||||
| NC score | 0.128155 (rank : 18) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3U4I7, Q8CAN2, Q9D630 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 homolog (EC 1.-.-.-). | |||||
|
PPOX_MOUSE
|
||||||
| NC score | 0.119316 (rank : 19) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51175, P97344, Q99M34 | Gene names | Ppox | |||
|
Domain Architecture |
|
|||||
| Description | Protoporphyrinogen oxidase (EC 1.3.3.4) (PPO). | |||||
|
FMO2_MOUSE
|
||||||
| NC score | 0.117796 (rank : 20) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K2I3, Q9QZF7 | Gene names | Fmo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2). | |||||
|
FMO3_MOUSE
|
||||||
| NC score | 0.114771 (rank : 21) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97501 | Gene names | Fmo3 | |||
|
Domain Architecture |
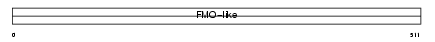
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3). | |||||
|
FMO3_HUMAN
|
||||||
| NC score | 0.110896 (rank : 22) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P31513, Q14854, Q8N5N5 | Gene names | FMO3 | |||
|
Domain Architecture |
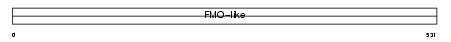
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 3 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 3) (FMO 3) (Dimethylaniline oxidase 3) (FMO form 2) (FMO II). | |||||
|
FMO2_HUMAN
|
||||||
| NC score | 0.109352 (rank : 23) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q99518, Q5EBX4, Q9BRX1 | Gene names | FMO2 | |||
|
Domain Architecture |
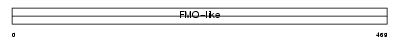
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 2 (EC 1.14.13.8) (Pulmonary flavin-containing monooxygenase 2) (FMO 2) (Dimethylaniline oxidase 2) (FMO 1B1). | |||||
|
CJ033_HUMAN
|
||||||
| NC score | 0.106997 (rank : 24) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N2H3, Q5TAA9, Q9BRQ1 | Gene names | C10orf33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable oxidoreductase C10orf33 (EC 1.-.-.-). | |||||
|
FMO1_HUMAN
|
||||||
| NC score | 0.102998 (rank : 25) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q01740, Q5QPT2, Q9UJC2 | Gene names | FMO1 | |||
|
Domain Architecture |
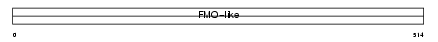
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Fetal hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
FMO1_MOUSE
|
||||||
| NC score | 0.095893 (rank : 26) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P50285 | Gene names | Fmo1, Fmo-1 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 1 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 1) (FMO 1) (Dimethylaniline oxidase 1). | |||||
|
FMO4_HUMAN
|
||||||
| NC score | 0.094711 (rank : 27) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P31512 | Gene names | FMO4, FMO2 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
FMO6_HUMAN
|
||||||
| NC score | 0.082677 (rank : 28) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60774 | Gene names | FMO6 | |||
|
Domain Architecture |
|
|||||
| Description | Putative dimethylaniline monooxygenase [N-oxide-forming] 6 (EC 1.14.13.8) (Flavin-containing monooxygenase 6) (FMO 6) (Dimethylaniline oxidase 6). | |||||
|
FMO4_MOUSE
|
||||||
| NC score | 0.081412 (rank : 29) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHG0 | Gene names | Fmo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 4 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 4) (FMO 4) (Dimethylaniline oxidase 4). | |||||
|
FMO5_HUMAN
|
||||||
| NC score | 0.078919 (rank : 30) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49326 | Gene names | FMO5 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 5) (FMO 5) (Dimethylaniline oxidase 5). | |||||
|
FMO5_MOUSE
|
||||||
| NC score | 0.074270 (rank : 31) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P97872 | Gene names | Fmo5 | |||
|
Domain Architecture |
|
|||||
| Description | Dimethylaniline monooxygenase [N-oxide-forming] 5 (EC 1.14.13.8) (Hepatic flavin-containing monooxygenase 5) (FMO 5) (Dimethylaniline oxidase 5). | |||||
|
PCYOX_MOUSE
|
||||||
| NC score | 0.069427 (rank : 32) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CQF9, Q3UHV6, Q69ZW0, Q8BZX1 | Gene names | Pcyox1, Kiaa0908 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prenylcysteine oxidase precursor (EC 1.8.3.5). | |||||
|
DLDH_HUMAN
|
||||||
| NC score | 0.068093 (rank : 33) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09622, Q14131, Q14167 | Gene names | DLD, GCSL, LAD, PHE3 | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) (Glycine cleavage system L protein). | |||||
|
DLDH_MOUSE
|
||||||
| NC score | 0.067983 (rank : 34) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O08749 | Gene names | Dld | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyl dehydrogenase, mitochondrial precursor (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase). | |||||
|
GSHR_MOUSE
|
||||||
| NC score | 0.045964 (rank : 35) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P47791, Q7TNC2, Q8BN97, Q8C9Z6 | Gene names | Gsr, Gr1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione reductase, mitochondrial precursor (EC 1.8.1.7) (GR) (GRase). | |||||
|
CB025_MOUSE
|
||||||
| NC score | 0.044673 (rank : 36) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LS1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf25 homolog, mitochondrial precursor. | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.040237 (rank : 37) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
CB025_HUMAN
|
||||||
| NC score | 0.038686 (rank : 38) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3L0, O95891 | Gene names | C2orf25, CL25022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C2orf25, mitochondrial precursor. | |||||
|
P3H3_HUMAN
|
||||||
| NC score | 0.035114 (rank : 39) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IVL6, Q13512, Q15740, Q66K32, Q6NX61, Q7L2T1 | Gene names | LEPREL2, P3H3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.034202 (rank : 40) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
CHDH_MOUSE
|
||||||
| NC score | 0.033027 (rank : 41) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJ64, Q8CHT7 | Gene names | Chdh | |||
|
Domain Architecture |

|
|||||
| Description | Choline dehydrogenase, mitochondrial precursor (EC 1.1.99.1) (CHD) (CDH). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.030881 (rank : 42) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
UN45A_HUMAN
|
||||||
| NC score | 0.027448 (rank : 43) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H3U1, Q7L3Y6, Q9H3U8, Q9NSE8, Q9NSE9 | Gene names | UNC45A, SMAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Smooth muscle cell-associated protein 1) (SMAP-1). | |||||
|
UN45A_MOUSE
|
||||||
| NC score | 0.026776 (rank : 44) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99KD5, Q3TKV6, Q8BFT3, Q8C157 | Gene names | Unc45a, Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UNC45 homolog A (UNC-45A) (Stromal membrane-associated protein 1) (Smap-1). | |||||
|
P3H3_MOUSE
|
||||||
| NC score | 0.025821 (rank : 45) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CG70, O88836 | Gene names | Leprel2, P3h3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prolyl 3-hydroxylase 3 precursor (EC 1.14.11.7) (Leprecan-like protein 2) (Protein B). | |||||
|
TCAL1_MOUSE
|
||||||
| NC score | 0.023771 (rank : 46) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q921P9 | Gene names | Tceal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 1 (TCEA-like protein 1) (Transcription elongation factor S-II protein-like 1). | |||||
|
FLVC2_MOUSE
|
||||||
| NC score | 0.018655 (rank : 47) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X85 | Gene names | Flvcr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Feline leukemia virus subgroup C receptor-related protein 2 (Calcium- chelate transporter) (CCT). | |||||
|
PHLA1_HUMAN
|
||||||
| NC score | 0.018126 (rank : 48) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WV24, Q15184, Q2TAN2, Q9NZ17 | Gene names | PHLDA1, PHRIP, TDAG51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family A member 1 (T-cell death- associated gene 51 protein) (Apoptosis-associated nuclear protein) (Proline- and histidine-rich protein) (Proline- and glutamine-rich protein) (PQ-rich protein). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.011537 (rank : 49) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.010814 (rank : 50) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||