Please be patient as the page loads
|
TSC2_MOUSE
|
||||||
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TSC2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991071 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
GRIPE_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 3) | NC score | 0.509368 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 4) | NC score | 0.507090 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 5) | NC score | 0.512755 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 6) | NC score | 0.411422 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 7) | NC score | 0.421090 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 8) | NC score | 0.415448 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 9) | NC score | 0.415387 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L1_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 10) | NC score | 0.410810 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 11) | NC score | 0.409614 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 12) | NC score | 0.423628 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
HD_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.054327 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.040525 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
VPS35_MOUSE
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.057976 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQH3, Q61123 | Gene names | Vps35, Mem3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (Maternal-embryonic 3). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.042011 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
FANCE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.100384 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HB96, Q4ZGH2 | Gene names | FANCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group E protein (Protein FACE). | |||||
|
PK1L1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.034995 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.020712 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.007015 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
STK23_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.018720 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0G2 | Gene names | Stk23, Mssk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 23 (EC 2.7.11.1) (Muscle-specific serine kinase 1) (MSSK-1). | |||||
|
VPS35_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.052638 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QK1, Q561W2, Q9H016, Q9H096, Q9H4P3, Q9H8J0, Q9NRS7, Q9NVG2, Q9NX80, Q9NZK2 | Gene names | VPS35, MEM3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (hVPS35) (Maternal-embryonic 3). | |||||
|
PARM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.038665 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.006359 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
CJ067_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.055096 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYJ2, Q5SWD4 | Gene names | C10orf67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf67. | |||||
|
FOXH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.010118 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |
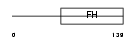
|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
GPR22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.013107 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99680, O14554 | Gene names | GPR22 | |||
|
Domain Architecture |
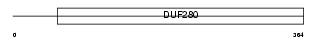
|
|||||
| Description | Probable G-protein coupled receptor 22. | |||||
|
CNTN6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.003134 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.009549 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
TCF19_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.021508 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KJ5, Q8BPA3, Q9CT93 | Gene names | Tcf19, Sc1 | |||
|
Domain Architecture |
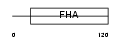
|
|||||
| Description | Transcription factor 19-like protein (SC1 protein homolog). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.012548 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.004294 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
KLC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.006734 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.014344 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
PER3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.008675 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PRR6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.019184 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXS4, Q5SX21, Q6PIA5 | Gene names | Prr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 6. | |||||
|
TEX10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.020587 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXF1, Q5T722, Q5T723, Q8NCN8, Q8TDY0 | Gene names | TEX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 10 protein. | |||||
|
CAN11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.004526 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
CTDP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.032565 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.000087 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DFNA5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.011748 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2D3 | Gene names | Dfna5, Dfna5h | |||
|
Domain Architecture |
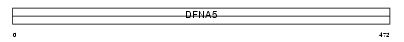
|
|||||
| Description | Non-syndromic hearing impairment protein 5 homolog. | |||||
|
ELF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.005867 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
FOXI1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.004833 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
HAIR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.008334 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.006521 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.010764 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.007972 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
TAF6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.014260 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49848 | Gene names | TAF6, TAF2E, TAFII70 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 6 (Transcription initiation factor TFIID 70 kDa subunit) (TAF(II)70) (TAFII-70) (TAFII- 80) (TAFII80). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.991071 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.512755 (rank : 3) | θ value | 1.2105e-12 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| NC score | 0.509368 (rank : 4) | θ value | 1.24977e-17 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| NC score | 0.507090 (rank : 5) | θ value | 2.13179e-17 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.423628 (rank : 6) | θ value | 1.38499e-08 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.421090 (rank : 7) | θ value | 2.13673e-09 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.415448 (rank : 8) | θ value | 3.64472e-09 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_MOUSE
|
||||||
| NC score | 0.415387 (rank : 9) | θ value | 3.64472e-09 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.411422 (rank : 10) | θ value | 2.13673e-09 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.410810 (rank : 11) | θ value | 1.06045e-08 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.409614 (rank : 12) | θ value | 1.06045e-08 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
FANCE_HUMAN
|
||||||
| NC score | 0.100384 (rank : 13) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HB96, Q4ZGH2 | Gene names | FANCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group E protein (Protein FACE). | |||||
|
VPS35_MOUSE
|
||||||
| NC score | 0.057976 (rank : 14) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQH3, Q61123 | Gene names | Vps35, Mem3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (Maternal-embryonic 3). | |||||
|
CJ067_HUMAN
|
||||||
| NC score | 0.055096 (rank : 15) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYJ2, Q5SWD4 | Gene names | C10orf67 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf67. | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.054327 (rank : 16) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
VPS35_HUMAN
|
||||||
| NC score | 0.052638 (rank : 17) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QK1, Q561W2, Q9H016, Q9H096, Q9H4P3, Q9H8J0, Q9NRS7, Q9NVG2, Q9NX80, Q9NZK2 | Gene names | VPS35, MEM3 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 35 (Vesicle protein sorting 35) (hVPS35) (Maternal-embryonic 3). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.042011 (rank : 18) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.040525 (rank : 19) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
PARM1_HUMAN
|
||||||
| NC score | 0.038665 (rank : 20) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWI2, Q96DV8, Q9Y4S1 | Gene names | ames=UNQ1879/PRO4322 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PK1L1_HUMAN
|
||||||
| NC score | 0.034995 (rank : 21) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDX9, Q6UWK1 | Gene names | PKD1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycystic kidney disease 1-like 1 protein (Polycystin-1L1). | |||||
|
CTDP1_MOUSE
|
||||||
| NC score | 0.032565 (rank : 22) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TSG2, Q7TSS7, Q9D4S8 | Gene names | Ctdp1, Fcp1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
TCF19_MOUSE
|
||||||
| NC score | 0.021508 (rank : 23) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KJ5, Q8BPA3, Q9CT93 | Gene names | Tcf19, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19-like protein (SC1 protein homolog). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.020712 (rank : 24) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
TEX10_HUMAN
|
||||||
| NC score | 0.020587 (rank : 25) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXF1, Q5T722, Q5T723, Q8NCN8, Q8TDY0 | Gene names | TEX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 10 protein. | |||||
|
PRR6_MOUSE
|
||||||
| NC score | 0.019184 (rank : 26) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CXS4, Q5SX21, Q6PIA5 | Gene names | Prr6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 6. | |||||
|
STK23_MOUSE
|
||||||
| NC score | 0.018720 (rank : 27) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0G2 | Gene names | Stk23, Mssk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 23 (EC 2.7.11.1) (Muscle-specific serine kinase 1) (MSSK-1). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.014344 (rank : 28) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
TAF6_HUMAN
|
||||||
| NC score | 0.014260 (rank : 29) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49848 | Gene names | TAF6, TAF2E, TAFII70 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 6 (Transcription initiation factor TFIID 70 kDa subunit) (TAF(II)70) (TAFII-70) (TAFII- 80) (TAFII80). | |||||
|
GPR22_HUMAN
|
||||||
| NC score | 0.013107 (rank : 30) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99680, O14554 | Gene names | GPR22 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 22. | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.012548 (rank : 31) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
DFNA5_MOUSE
|
||||||
| NC score | 0.011748 (rank : 32) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2D3 | Gene names | Dfna5, Dfna5h | |||
|
Domain Architecture |

|
|||||
| Description | Non-syndromic hearing impairment protein 5 homolog. | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.010764 (rank : 33) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
FOXH1_MOUSE
|
||||||
| NC score | 0.010118 (rank : 34) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88621, Q9QZL5, Q9R241 | Gene names | Foxh1, Fast1, Fast2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.009549 (rank : 35) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.008675 (rank : 36) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
HAIR_HUMAN
|
||||||
| NC score | 0.008334 (rank : 37) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43593, Q96H33, Q9NPE1 | Gene names | HR | |||
|
Domain Architecture |

|
|||||
| Description | Protein hairless. | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.007972 (rank : 38) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.007015 (rank : 39) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
KLC1_HUMAN
|
||||||
| NC score | 0.006734 (rank : 40) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07866, Q7RTM2, Q7RTM3, Q7RTM5, Q7RTP9, Q7RTQ5, Q86SF5, Q86TF5, Q86V74, Q86V75, Q86V76, Q86V77, Q86V78, Q86V79, Q96H62 | Gene names | KNS2, KLC, KLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin light chain 1 (KLC 1). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.006521 (rank : 41) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.006359 (rank : 42) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
ELF1_MOUSE
|
||||||
| NC score | 0.005867 (rank : 43) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60775 | Gene names | Elf1 | |||
|
Domain Architecture |

|
|||||
| Description | ETS-related transcription factor Elf-1 (E74-like factor 1). | |||||
|
FOXI1_HUMAN
|
||||||
| NC score | 0.004833 (rank : 44) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
CAN11_HUMAN
|
||||||
| NC score | 0.004526 (rank : 45) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UMQ6, Q8N4R5 | Gene names | CAPN11 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-11 (EC 3.4.22.-) (Calcium-activated neutral proteinase 11) (CANP 11). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.004294 (rank : 46) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
CNTN6_MOUSE
|
||||||
| NC score | 0.003134 (rank : 47) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.000087 (rank : 48) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||