Please be patient as the page loads
|
GRIPE_HUMAN
|
||||||
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GRIPE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992795 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 3) | NC score | 0.527405 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 4) | NC score | 0.509368 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 5) | NC score | 0.542469 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 6) | NC score | 0.463539 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_MOUSE
|
||||||
| θ value | 4.16044e-13 (rank : 7) | NC score | 0.462192 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 8) | NC score | 0.458304 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 9) | NC score | 0.456969 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 10) | NC score | 0.453460 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 11) | NC score | 0.458431 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SIPA1_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 12) | NC score | 0.446612 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
K1219_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 13) | NC score | 0.236948 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
K1219_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 14) | NC score | 0.228716 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BQZ4, Q80TH5, Q8BQN1 | Gene names | Kiaa1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.036046 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.041172 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.078456 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.030780 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.020062 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
FOLH1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.038742 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35409, Q9DCC2 | Gene names | Folh1, Mopsm, Naalad1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate carboxypeptidase 2 (EC 3.4.17.21) (Glutamate carboxypeptidase II) (Membrane glutamate carboxypeptidase) (mGCP) (N- acetylated-alpha-linked acidic dipeptidase I) (NAALADase I) (Pteroylpoly-gamma-glutamate carboxypeptidase) (Folylpoly-gamma- glutamate carboxypeptidase) (FGCP) (Folate hydrolase 1) (Prostate- specific membrane antigen homolog). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.019727 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.030371 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
FOLH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.035399 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04609, O43748, Q16305, Q9NP15, Q9NYE2, Q9P1P8 | Gene names | FOLH1, FOLH, NAALAD1, PSM, PSMA | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate carboxypeptidase 2 (EC 3.4.17.21) (Glutamate carboxypeptidase II) (Membrane glutamate carboxypeptidase) (mGCP) (N- acetylated-alpha-linked acidic dipeptidase I) (NAALADase I) (Pteroylpoly-gamma-glutamate carboxypeptidase) (Folylpoly-gamma- glutamate carboxypeptidase) (FGCP) (Folate hydrolase 1) (Prostate- specific membrane antigen) (PSMA) (PSM). | |||||
|
RP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.022713 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.013516 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.007850 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.020391 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.011503 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
ZNF18_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.001879 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17022, Q5QHQ3, Q8IYC4, Q8NAH6 | Gene names | ZNF18, HDSG1, KOX11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein KOX11) (Heart development- specific gene 1 protein). | |||||
|
A20A1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.017725 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 698 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TYW2, Q9H0H6 | Gene names | ANKRD20A1, ANKRD20A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A1. | |||||
|
A20A3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.017893 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VUR7 | Gene names | ANKRD20A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A3. | |||||
|
A20A4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.017716 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SQ80 | Gene names | ANKRD20A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A4. | |||||
|
OTU7A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.031722 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.031489 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.019091 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
A20A2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.017067 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q4UJ75 | Gene names | ANKRD20A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A2. | |||||
|
CH3L1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.018545 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36222, P30923, Q8IVA4, Q96HI7 | Gene names | CHI3L1 | |||
|
Domain Architecture |
|
|||||
| Description | Chitinase-3-like protein 1 precursor (Cartilage glycoprotein 39) (GP- 39) (39 kDa synovial protein) (HCgp-39) (YKL-40). | |||||
|
DBR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.020219 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923B1, Q8C1T9, Q8C7J7, Q99MT1 | Gene names | Dbr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lariat debranching enzyme (EC 3.1.-.-). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.023768 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.023570 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
ANR15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.008826 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
ARFG3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.011742 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |
|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.020047 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.011194 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
REST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.007698 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
T2FB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.020798 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0A0 | Gene names | Gtf2f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIF subunit beta (EC 3.6.1.-) (ATP- dependent helicase GTF2F2) (TFIIF-beta). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.006506 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.013105 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.016299 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
KI2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.004987 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.007669 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TOX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.010688 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TRAP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.010602 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.007447 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
APC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.023909 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
DNL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.009411 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.002122 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
GP152_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.007392 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
MAGAC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.004113 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.010107 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
ROBO2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.002591 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
ROBO2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.002620 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.009624 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.016281 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.006728 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.012396 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.012992 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TRPS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.007745 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
TRS85_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.025945 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L5, Q9H0L2 | Gene names | KIAA1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TRS85 homolog. | |||||
|
GRIPE_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| NC score | 0.992795 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.542469 (rank : 3) | θ value | 4.91457e-14 (rank : 5) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.527405 (rank : 4) | θ value | 4.58923e-20 (rank : 3) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.509368 (rank : 5) | θ value | 1.24977e-17 (rank : 4) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.463539 (rank : 6) | θ value | 3.18553e-13 (rank : 6) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_MOUSE
|
||||||
| NC score | 0.462192 (rank : 7) | θ value | 4.16044e-13 (rank : 7) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.458431 (rank : 8) | θ value | 1.133e-10 (rank : 11) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.458304 (rank : 9) | θ value | 7.09661e-13 (rank : 8) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.456969 (rank : 10) | θ value | 7.09661e-13 (rank : 9) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.453460 (rank : 11) | θ value | 2.0648e-12 (rank : 10) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.446612 (rank : 12) | θ value | 9.59137e-10 (rank : 12) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
K1219_HUMAN
|
||||||
| NC score | 0.236948 (rank : 13) | θ value | 1.99992e-07 (rank : 13) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86X10, Q5TG31, Q8N3D1, Q8WWC0, Q9H3X8, Q9UJR1, Q9ULK1, Q9Y3G9 | Gene names | KIAA1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
K1219_MOUSE
|
||||||
| NC score | 0.228716 (rank : 14) | θ value | 1.69304e-06 (rank : 14) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BQZ4, Q80TH5, Q8BQN1 | Gene names | Kiaa1219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1219. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.078456 (rank : 15) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.041172 (rank : 16) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
FOLH1_MOUSE
|
||||||
| NC score | 0.038742 (rank : 17) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35409, Q9DCC2 | Gene names | Folh1, Mopsm, Naalad1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate carboxypeptidase 2 (EC 3.4.17.21) (Glutamate carboxypeptidase II) (Membrane glutamate carboxypeptidase) (mGCP) (N- acetylated-alpha-linked acidic dipeptidase I) (NAALADase I) (Pteroylpoly-gamma-glutamate carboxypeptidase) (Folylpoly-gamma- glutamate carboxypeptidase) (FGCP) (Folate hydrolase 1) (Prostate- specific membrane antigen homolog). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.036046 (rank : 18) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
FOLH1_HUMAN
|
||||||
| NC score | 0.035399 (rank : 19) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04609, O43748, Q16305, Q9NP15, Q9NYE2, Q9P1P8 | Gene names | FOLH1, FOLH, NAALAD1, PSM, PSMA | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate carboxypeptidase 2 (EC 3.4.17.21) (Glutamate carboxypeptidase II) (Membrane glutamate carboxypeptidase) (mGCP) (N- acetylated-alpha-linked acidic dipeptidase I) (NAALADase I) (Pteroylpoly-gamma-glutamate carboxypeptidase) (Folylpoly-gamma- glutamate carboxypeptidase) (FGCP) (Folate hydrolase 1) (Prostate- specific membrane antigen) (PSMA) (PSM). | |||||
|
OTU7A_HUMAN
|
||||||
| NC score | 0.031722 (rank : 20) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TE49, Q8IWK5 | Gene names | OTUD7A, C15orf16, OTUD7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
OTU7A_MOUSE
|
||||||
| NC score | 0.031489 (rank : 21) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R554 | Gene names | Otud7a, Otud7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | OTU domain-containing protein 7A (EC 3.-.-.-) (Zinc finger protein Cezanne 2). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.030780 (rank : 22) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.030371 (rank : 23) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
TRS85_HUMAN
|
||||||
| NC score | 0.025945 (rank : 24) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2L5, Q9H0L2 | Gene names | KIAA1012 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TRS85 homolog. | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.023909 (rank : 25) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.023768 (rank : 26) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.023570 (rank : 27) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
RP1_HUMAN
|
||||||
| NC score | 0.022713 (rank : 28) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56715 | Gene names | RP1, ORP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein) (Retinitis pigmentosa 1 protein). | |||||
|
T2FB_MOUSE
|
||||||
| NC score | 0.020798 (rank : 29) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0A0 | Gene names | Gtf2f2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor IIF subunit beta (EC 3.6.1.-) (ATP- dependent helicase GTF2F2) (TFIIF-beta). | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.020391 (rank : 30) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
DBR1_MOUSE
|
||||||
| NC score | 0.020219 (rank : 31) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923B1, Q8C1T9, Q8C7J7, Q99MT1 | Gene names | Dbr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lariat debranching enzyme (EC 3.1.-.-). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.020062 (rank : 32) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.020047 (rank : 33) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.019727 (rank : 34) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.019091 (rank : 35) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
CH3L1_HUMAN
|
||||||
| NC score | 0.018545 (rank : 36) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P36222, P30923, Q8IVA4, Q96HI7 | Gene names | CHI3L1 | |||
|
Domain Architecture |
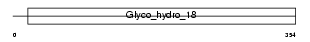
|
|||||
| Description | Chitinase-3-like protein 1 precursor (Cartilage glycoprotein 39) (GP- 39) (39 kDa synovial protein) (HCgp-39) (YKL-40). | |||||
|
A20A3_HUMAN
|
||||||
| NC score | 0.017893 (rank : 37) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5VUR7 | Gene names | ANKRD20A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A3. | |||||
|
A20A1_HUMAN
|
||||||
| NC score | 0.017725 (rank : 38) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 698 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5TYW2, Q9H0H6 | Gene names | ANKRD20A1, ANKRD20A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A1. | |||||
|
A20A4_HUMAN
|
||||||
| NC score | 0.017716 (rank : 39) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5SQ80 | Gene names | ANKRD20A4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A4. | |||||
|
A20A2_HUMAN
|
||||||
| NC score | 0.017067 (rank : 40) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q4UJ75 | Gene names | ANKRD20A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 20A2. | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.016299 (rank : 41) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.016281 (rank : 42) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.013516 (rank : 43) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.013105 (rank : 44) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.012992 (rank : 45) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.012396 (rank : 46) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
ARFG3_HUMAN
|
||||||
| NC score | 0.011742 (rank : 47) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NP61, Q9BSC6, Q9H9J0, Q9NT10, Q9NUP5, Q9Y4V3, Q9Y4V4 | Gene names | ARFGAP3, ARFGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor GTPase-activating protein 3 (ARF GAP 3). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.011503 (rank : 48) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.011194 (rank : 49) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.010688 (rank : 50) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TRAP1_HUMAN
|
||||||
| NC score | 0.010602 (rank : 51) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12931, O43642, O75235, Q9UHL5 | Gene names | TRAP1, HSP75 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock protein 75 kDa, mitochondrial precursor (HSP 75) (Tumor necrosis factor type 1 receptor-associated protein) (TRAP-1) (TNFR- associated protein 1). | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.010107 (rank : 52) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.009624 (rank : 53) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
DNL3_HUMAN
|
||||||
| NC score | 0.009411 (rank : 54) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49916, Q16714 | Gene names | LIG3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 3 (EC 6.5.1.1) (DNA ligase III) (Polydeoxyribonucleotide synthase [ATP] 3). | |||||
|
ANR15_HUMAN
|
||||||
| NC score | 0.008826 (rank : 55) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14678, Q5W0W0, Q8IY65, Q8WX74 | Gene names | ANKRD15, KANK, KIAA0172 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 15 (Kidney ankyrin repeat- containing protein). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.007850 (rank : 56) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
TRPS1_HUMAN
|
||||||
| NC score | 0.007745 (rank : 57) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHF7, Q9NWE1, Q9UHH6 | Gene names | TRPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger transcription factor Trps1 (Zinc finger protein GC79) (Tricho-rhino-phalangeal syndrome type I protein). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.007698 (rank : 58) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.007669 (rank : 59) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.007447 (rank : 60) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.007392 (rank : 61) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.006728 (rank : 62) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.006506 (rank : 63) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
KI2L1_HUMAN
|
||||||
| NC score | 0.004987 (rank : 64) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
MAGAC_HUMAN
|
||||||
| NC score | 0.004113 (rank : 65) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43365, Q9NSD3 | Gene names | MAGEA12, MAGE12 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 12 (MAGE-12 antigen) (MAGE12F). | |||||
|
ROBO2_MOUSE
|
||||||
| NC score | 0.002620 (rank : 66) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TPD3, Q8BJ59 | Gene names | Robo2, Kiaa1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
ROBO2_HUMAN
|
||||||
| NC score | 0.002591 (rank : 67) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCK4, O43608 | Gene names | ROBO2, KIAA1568 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 2 precursor. | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.002122 (rank : 68) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
ZNF18_HUMAN
|
||||||
| NC score | 0.001879 (rank : 69) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 69 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17022, Q5QHQ3, Q8IYC4, Q8NAH6 | Gene names | ZNF18, HDSG1, KOX11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein KOX11) (Heart development- specific gene 1 protein). | |||||