Please be patient as the page loads
|
SIPA1_MOUSE
|
||||||
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SIPA1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.976385 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 2.03516e-177 (rank : 3) | NC score | 0.937806 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L1_HUMAN
|
||||||
| θ value | 9.45368e-175 (rank : 4) | NC score | 0.940008 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L2_MOUSE
|
||||||
| θ value | 6.77497e-173 (rank : 5) | NC score | 0.942993 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_HUMAN
|
||||||
| θ value | 2.84643e-171 (rank : 6) | NC score | 0.941434 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L3_HUMAN
|
||||||
| θ value | 7.25522e-167 (rank : 7) | NC score | 0.925413 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
RGP2_HUMAN
|
||||||
| θ value | 6.77864e-56 (rank : 8) | NC score | 0.894291 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 9) | NC score | 0.458431 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 10) | NC score | 0.446232 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 11) | NC score | 0.443297 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 12) | NC score | 0.423628 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.071796 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.069136 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
PERI_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.030853 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.014072 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.013780 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
PERI_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.030518 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |
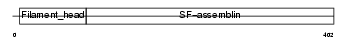
|
|||||
| Description | Peripherin. | |||||
|
STXB4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.060997 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.054860 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.035716 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.052280 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.061246 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.056530 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.055076 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SKIL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.054159 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |
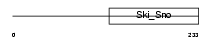
|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.054652 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.058056 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.056685 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.031093 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ATD3B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.058364 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.012606 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
SGOL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.039454 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.052473 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
ATAD3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.057992 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
ATD3A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.055738 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.018810 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
NAF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.066811 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.059009 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 40) | NC score | 0.031006 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SDCB2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 41) | NC score | 0.039480 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H190, O95892, Q5W0X1, Q9BZ42, Q9H567, Q9NRY8 | Gene names | SDCBP2, SITAC18 | |||
|
Domain Architecture |
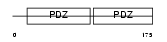
|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.048787 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
HOOK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 43) | NC score | 0.055334 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
IKBL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.025269 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |
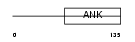
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
CAC1H_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.015649 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.047926 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.007263 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
HOOK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 48) | NC score | 0.062095 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
MYO1F_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.022384 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00160, Q8WWN7 | Gene names | MYO1F | |||
|
Domain Architecture |

|
|||||
| Description | Myosin If (Myosin-IE). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.064031 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.048935 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 52) | NC score | 0.028941 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
RUFY1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 53) | NC score | 0.067479 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.065418 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.045754 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
CTGE2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.039769 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
MAF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 57) | NC score | 0.020732 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_MOUSE
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.020315 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.052811 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.068296 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
TPM2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.048222 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TRI39_MOUSE
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.013661 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9ESN2, Q8BPR5, Q8K0F7 | Gene names | Trim39, Rnf23, Tfp | |||
|
Domain Architecture |
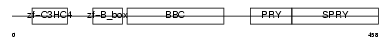
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.052325 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
AMOL2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.042506 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.051815 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CP250_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.056820 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DESM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.027450 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |
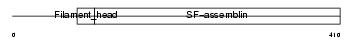
|
|||||
| Description | Desmin. | |||||
|
DESM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.027249 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P31001 | Gene names | Des | |||
|
Domain Architecture |
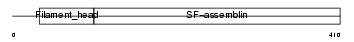
|
|||||
| Description | Desmin. | |||||
|
EP15_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.037358 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.050091 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.051909 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 72) | NC score | 0.051651 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 73) | NC score | 0.053450 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 74) | NC score | 0.048634 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.085214 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.085624 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.055375 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.050491 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.033295 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.045873 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
REST_HUMAN
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.054589 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TRI45_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.009484 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PFY8, Q8BVT5 | Gene names | Trim45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.024024 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.053586 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.053626 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CO8A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.004049 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.024603 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
DMPK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.008256 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
DP13A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.027225 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.059005 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
KIF3C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.014601 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |
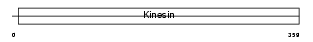
|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.022688 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.007116 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MFAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.028401 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CQU1, Q3TU29, Q8CCL1, Q9CSJ5 | Gene names | Mfap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 1. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.009104 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.051678 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.051462 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.058408 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
REST_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.055719 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RUFY1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.061424 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
SELO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.030287 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BVL4, Q8WUI0 | Gene names | SELO | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein O. | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.025135 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
XKR5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.011973 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
XRN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.018509 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0D6, Q3L8N4, Q6KGZ9, Q9BQL1, Q9NTW0, Q9NXS6, Q9UL53 | Gene names | XRN2 | |||
|
Domain Architecture |
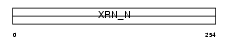
|
|||||
| Description | 5'-3' exoribonuclease 2 (EC 3.1.13.-) (DHM1-like protein) (DHP protein). | |||||
|
ANR18_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.031973 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
APBA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.082460 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |
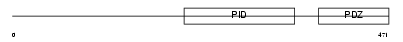
|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.003887 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.049022 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
DEMA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.009044 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08495, Q13215, Q9BRE3 | Gene names | EPB49, DMT | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
DP13A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.027631 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.008177 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.050912 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
K1H7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.019531 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O76014 | Gene names | KRTHA7, HHA7, HKA7 | |||
|
Domain Architecture |
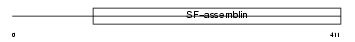
|
|||||
| Description | Keratin, type I cuticular Ha7 (Hair keratin, type I Ha7). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.029131 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.045352 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.048131 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.018944 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.052196 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.058255 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.008263 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.005342 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TPM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.046589 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
TRI39_HUMAN
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | 0.012331 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |
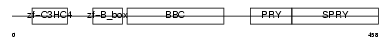
|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.001545 (rank : 184) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.010455 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CENPQ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.029380 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CPQ5, Q8C2G4, Q8VEF8, Q9CX70 | Gene names | Cenpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
CING_HUMAN
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.050585 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
ES8L1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.007467 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.050739 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.014013 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.056831 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.003253 (rank : 183) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
STON1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.007889 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.026755 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TPM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.045267 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.045082 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.011542 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ADDB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.006405 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
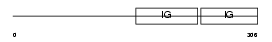
CEAM8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.001403 (rank : 185) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31997, O60399, Q16574 | Gene names | CEACAM8, CGM6 | |||
|
Domain Architecture |
|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor (Carcinoembryonic antigen CGM6) (Nonspecific cross-reacting antigen NCA-95) (CD67 antigen) (CD66b antigen). | |||||
|
DCJ14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.005157 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.049285 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.008971 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.044725 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
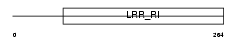
LUM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.005087 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51885, Q3TP25, Q99JZ3, Q9CXK0 | Gene names | Lum, Lcn, Ldc | |||
|
Domain Architecture |
|
|||||
| Description | Lumican precursor (Keratan sulfate proteoglycan lumican) (KSPG lumican). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.045274 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.044803 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.043384 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.043657 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
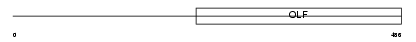
MYOC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.016714 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O70624, O70289 | Gene names | Myoc, Tigr | |||
|
Domain Architecture |
|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
PAXI_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.007106 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI36, Q3TB62, Q3TZQ6, Q8VI37 | Gene names | Pxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paxillin. | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.022419 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.022791 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
ST18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.022994 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.035859 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.062073 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.064050 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.016035 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZMYM3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.009031 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.068513 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
APBA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.065041 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
CCDC6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.062611 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.051834 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050194 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.050532 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.050245 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.052449 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.053144 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052688 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.055461 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.055811 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.057434 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
HOOK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.052567 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.052700 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.052185 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.050446 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.058684 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.053183 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
RUFY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.052498 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
RUFY3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052994 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.060047 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.072369 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.063202 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
TPR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.056662 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
TUFT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.052907 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
VGF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.050567 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 158 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SIPA1_HUMAN
|
||||||
| NC score | 0.976385 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q96FS4, O14518, O60484, O60618 | Gene names | SIPA1, SPA1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1) (p130 SPA-1). | |||||
|
SI1L2_MOUSE
|
||||||
| NC score | 0.942993 (rank : 3) | θ value | 6.77497e-173 (rank : 5) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80TE4, Q6PDY1 | Gene names | Sipa1l2, Kiaa0545 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L2_HUMAN
|
||||||
| NC score | 0.941434 (rank : 4) | θ value | 2.84643e-171 (rank : 6) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9P2F8, Q5VXR7, Q5VXR8, Q641Q4, Q8NA38, Q96DZ3, Q9H9F6 | Gene names | SIPA1L2, KIAA1389 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 2. | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.940008 (rank : 5) | θ value | 9.45368e-175 (rank : 4) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.937806 (rank : 6) | θ value | 2.03516e-177 (rank : 3) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
SI1L3_HUMAN
|
||||||
| NC score | 0.925413 (rank : 7) | θ value | 7.25522e-167 (rank : 7) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O60292, Q2TV87 | Gene names | SIPA1L3, KIAA0545, SPAL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 3 (SPA-1-like protein 3). | |||||
|
RGP2_HUMAN
|
||||||
| NC score | 0.894291 (rank : 8) | θ value | 6.77864e-56 (rank : 8) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P47736 | Gene names | RAP1GAP, RAP1GA1 | |||
|
Domain Architecture |

|
|||||
| Description | Rap1 GTPase-activating protein 1 (Rap1GAP). | |||||
|
GRIPE_HUMAN
|
||||||
| NC score | 0.458431 (rank : 9) | θ value | 1.133e-10 (rank : 9) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6GYQ0, O94960, Q6GYP9, Q6ZT23, Q86YF3, Q86YF5, Q8ND69 | Gene names | GARNL1, KIAA0884, TULIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating Rap/Ran-GAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
GRIPE_MOUSE
|
||||||
| NC score | 0.446232 (rank : 10) | θ value | 9.59137e-10 (rank : 10) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6GYP7, Q6GYP6, Q6ZQ28, Q8BNJ5, Q8C9G8, Q8CIW4, Q9JMC4 | Gene names | Garnl1, Kiaa0884, Tulip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTPase-activating RapGAP domain-like 1 (GAP-related-interacting partner to E12) (GRIPE) (Tuberin-like protein 1). | |||||
|
TSC2_HUMAN
|
||||||
| NC score | 0.443297 (rank : 11) | θ value | 1.63604e-09 (rank : 11) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P49815, O75275, Q8TAZ1 | Gene names | TSC2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 protein). | |||||
|
TSC2_MOUSE
|
||||||
| NC score | 0.423628 (rank : 12) | θ value | 1.38499e-08 (rank : 12) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61037, P97723, P97724, P97725, P97727, Q61007, Q61008, Q9WUF6 | Gene names | Tsc2 | |||
|
Domain Architecture |

|
|||||
| Description | Tuberin (Tuberous sclerosis 2 homolog protein). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.085624 (rank : 13) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.085214 (rank : 14) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA3_MOUSE
|
||||||
| NC score | 0.082460 (rank : 15) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.072369 (rank : 16) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.071796 (rank : 17) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.069136 (rank : 18) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.068513 (rank : 19) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.068296 (rank : 20) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
RUFY1_HUMAN
|
||||||
| NC score | 0.067479 (rank : 21) | θ value | 1.38821 (rank : 53) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 848 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q96T51, Q59FF3, Q71S93, Q9H6I3 | Gene names | RUFY1, RABIP4, ZFYVE12 | |||
|
Domain Architecture |

|
|||||
| Description | RUN and FYVE domain-containing protein 1 (FYVE-finger protein EIP1) (Zinc finger FYVE domain-containing protein 12) (La-binding protein 1) (Rab4-interacting protein). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.066811 (rank : 22) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.065418 (rank : 23) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.065041 (rank : 24) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.064050 (rank : 25) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.064031 (rank : 26) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.063202 (rank : 27) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
CCDC6_HUMAN
|
||||||
| NC score | 0.062611 (rank : 28) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q16204, Q15250 | Gene names | CCDC6, D10S170 | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil domain-containing protein 6 (H4 protein) (Papillary thyroid carcinoma-encoded protein). | |||||
|
HOOK2_MOUSE
|
||||||
| NC score | 0.062095 (rank : 29) | θ value | 1.38821 (rank : 48) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7TMK6, Q8BY47, Q8R347, Q8VCN4, Q99LU2 | Gene names | Hook2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2. | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.062073 (rank : 30) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
RUFY1_MOUSE
|
||||||
| NC score | 0.061424 (rank : 31) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q8BIJ7, Q8BKQ4, Q8BL21, Q9EPM6 | Gene names | Rufy1, Rabip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 1 (Rab4-interacting protein). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.061246 (rank : 32) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
STXB4_HUMAN
|
||||||
| NC score | 0.060997 (rank : 33) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 616 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q6ZWJ1, Q8IVZ5 | Gene names | STXBP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaxin-binding protein 4 (Syntaxin 4-interacting protein) (Synip). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.060047 (rank : 34) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.059009 (rank : 35) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.059005 (rank : 36) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.058684 (rank : 37) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.058408 (rank : 38) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
ATD3B_HUMAN
|
||||||
| NC score | 0.058364 (rank : 39) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5T9A4, Q6ZRB5, Q9BUK4, Q9ULE7 | Gene names | ATAD3B, KIAA1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3B. | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.058255 (rank : 40) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.058056 (rank : 41) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
ATAD3_MOUSE
|
||||||
| NC score | 0.057992 (rank : 42) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q925I1, Q3UE74, Q69ZM7, Q8C6C6 | Gene names | Atad3, Atad3a, Kiaa1273 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3 (AAA-ATPase TOB3). | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.057434 (rank : 43) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.056831 (rank : 44) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.056820 (rank : 45) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.056685 (rank : 46) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.056662 (rank : 47) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.056530 (rank : 48) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.055811 (rank : 49) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
ATD3A_HUMAN
|
||||||
| NC score | 0.055738 (rank : 50) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NVI7, Q8N275, Q96A50 | Gene names | ATAD3A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATPase family AAA domain-containing protein 3A. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.055719 (rank : 51) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.055461 (rank : 52) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.055375 (rank : 53) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
HOOK1_MOUSE
|
||||||
| NC score | 0.055334 (rank : 54) | θ value | 1.06291 (rank : 43) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q8BIL5, Q8BIZ2, Q8K0P1, Q8K454, Q9CTN6 | Gene names | Hook1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.055076 (rank : 55) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.054860 (rank : 56) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.054652 (rank : 57) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.054589 (rank : 58) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SKIL_HUMAN
|
||||||
| NC score | 0.054159 (rank : 59) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.053626 (rank : 60) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.053586 (rank : 61) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.053450 (rank : 62) | θ value | 2.36792 (rank : 73) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.053183 (rank : 63) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.053144 (rank : 64) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
RUFY3_MOUSE
|
||||||
| NC score | 0.052994 (rank : 65) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9D394, Q3U348, Q3UF96, Q6PE64, Q8VD10 | Gene names | Rufy3, D5Bwg0860e, Ripx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
TUFT1_HUMAN
|
||||||
| NC score | 0.052907 (rank : 66) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 686 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9NNX1, Q5T384, Q5T385, Q9BU28, Q9H5L1 | Gene names | TUFT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tuftelin. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.052811 (rank : 67) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.052700 (rank : 68) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.052688 (rank : 69) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
HOOK3_HUMAN
|
||||||
| NC score | 0.052567 (rank : 70) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q86VS8, Q8NBH0, Q9BY13 | Gene names | HOOK3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3 (hHK3). | |||||
|
RUFY3_HUMAN
|
||||||
| NC score | 0.052498 (rank : 71) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q7L099, O94948, Q9UI00 | Gene names | RUFY3, KIAA0871 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein RUFY3 (Rap2-interacting protein x) (RIPx). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.052473 (rank : 72) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.052449 (rank : 73) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.052325 (rank : 74) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.052280 (rank : 75) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.052196 (rank : 76) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.052185 (rank : 77) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.051909 (rank : 78) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.051834 (rank : 79) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.051815 (rank : 80) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.051678 (rank : 81) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.051651 (rank : 82) | θ value | 2.36792 (rank : 72) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.051462 (rank : 83) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.050912 (rank : 84) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.050739 (rank : 85) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.050585 (rank : 86) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
VGF_HUMAN
|
||||||
| NC score | 0.050567 (rank : 87) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.050532 (rank : 88) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.050491 (rank : 89) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.050446 (rank : 90) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.050245 (rank : 91) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.050194 (rank : 92) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.050091 (rank : 93) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.049285 (rank : 94) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.049022 (rank : 95) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.048935 (rank : 96) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.048787 (rank : 97) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.048634 (rank : 98) | θ value | 2.36792 (rank : 74) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
TPM2_MOUSE
|
||||||
| NC score | 0.048222 (rank : 99) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P58774, P02560, P46901 | Gene names | Tpm2, Tpm-2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.048131 (rank : 100) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.047926 (rank : 101) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
TPM2_HUMAN
|
||||||
| NC score | 0.046589 (rank : 102) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 677 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P07951, P06468, Q13894, Q53FM4, Q5TCU4, Q5TCU7, Q9UH67 | Gene names | TPM2, TMSB | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin beta chain (Tropomyosin 2) (Beta-tropomyosin). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.045873 (rank : 103) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.045754 (rank : 104) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.045352 (rank : 105) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.045274 (rank : 106) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
TPM1_HUMAN
|
||||||
| NC score | 0.045267 (rank : 107) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 716 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P09493, P09494, P10469, Q86W64, Q96IK2, Q9UCY9 | Gene names | TPM1, TMSA | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
TPM1_MOUSE
|
||||||
| NC score | 0.045082 (rank : 108) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 729 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P58771, P02558, P19354, P46902, P99034 | Gene names | Tpm1, Tpm-1, Tpma | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin 1 alpha chain (Alpha-tropomyosin). | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.044803 (rank : 109) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.044725 (rank : 110) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.043657 (rank : 111) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH4_HUMAN
|
||||||
| NC score | 0.043384 (rank : 112) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1377 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Y623 | Gene names | MYH4 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal) (Myosin heavy chain IIb) (MyHC-IIb). | |||||
|
AMOL2_MOUSE
|
||||||
| NC score | 0.042506 (rank : 113) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8K371, Q3TPM1, Q7TPE4, Q8BP84, Q8BS08, Q9QUS0 | Gene names | Amotl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin-like protein 2. | |||||
|
CTGE2_HUMAN
|
||||||
| NC score | 0.039769 (rank : 114) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 594 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q96RT6 | Gene names | CTAGE1, CTAGE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-2. | |||||
|
SDCB2_HUMAN
|
||||||
| NC score | 0.039480 (rank : 115) | θ value | 0.813845 (rank : 41) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H190, O95892, Q5W0X1, Q9BZ42, Q9H567, Q9NRY8 | Gene names | SDCBP2, SITAC18 | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
SGOL1_MOUSE
|
||||||
| NC score | 0.039454 (rank : 116) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXH7, Q3U4K4, Q588H1, Q8BKW2 | Gene names | Sgol1, Sgo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 1. | |||||
|
EP15_MOUSE
|
||||||
| NC score | 0.037358 (rank : 117) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | P42567 | Gene names | Eps15 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.035859 (rank : 118) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.035716 (rank : 119) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.033295 (rank : 120) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
ANR18_HUMAN
|
||||||
| NC score | 0.031973 (rank : 121) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1106 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q8IVF6, Q7Z468 | Gene names | ANKRD18A, KIAA2015 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 18A. | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.031093 (rank : 122) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.031006 (rank : 123) | θ value | 0.813845 (rank : 40) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
PERI_MOUSE
|
||||||
| NC score | 0.030853 (rank : 124) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P15331, O35688, O35689 | Gene names | Prph, Prph1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
PERI_HUMAN
|
||||||
| NC score | 0.030518 (rank : 125) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P41219, Q8N577 | Gene names | PRPH | |||
|
Domain Architecture |

|
|||||
| Description | Peripherin. | |||||
|
SELO_HUMAN
|
||||||
| NC score | 0.030287 (rank : 126) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BVL4, Q8WUI0 | Gene names | SELO | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein O. | |||||
|
CENPQ_MOUSE
|
||||||
| NC score | 0.029380 (rank : 127) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CPQ5, Q8C2G4, Q8VEF8, Q9CX70 | Gene names | Cenpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein Q (CENP-Q). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.029131 (rank : 128) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.028941 (rank : 129) | θ value | 1.38821 (rank : 52) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
MFAP1_MOUSE
|
||||||
| NC score | 0.028401 (rank : 130) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9CQU1, Q3TU29, Q8CCL1, Q9CSJ5 | Gene names | Mfap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microfibrillar-associated protein 1. | |||||
|
DP13A_MOUSE
|
||||||
| NC score | 0.027631 (rank : 131) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3H0, Q3UJP7, Q69ZJ9, Q8BWZ8, Q8VCJ8, Q9CUW4 | Gene names | Appl1, Dip13a, Kiaa1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
DESM_HUMAN
|
||||||
| NC score | 0.027450 (rank : 132) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P17661, Q15787, Q8IZR1, Q8IZR6, Q8NES2, Q8NEU6, Q8TAC4, Q8TCX2, Q8TD99, Q9UHN5, Q9UJ80 | Gene names | DES | |||
|
Domain Architecture |

|
|||||
| Description | Desmin. | |||||
|
DESM_MOUSE
|
||||||
| NC score | 0.027249 (rank : 133) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P31001 | Gene names | Des | |||
|
Domain Architecture |

|
|||||
| Description | Desmin. | |||||
|
DP13A_HUMAN
|
||||||
| NC score | 0.027225 (rank : 134) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UKG1, Q9P2B9 | Gene names | APPL1, APPL, DIP13A, KIAA1428 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DCC-interacting protein 13 alpha (Dip13 alpha) (Adapter protein containing PH domain, PTB domain and leucine zipper motif 1). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.026755 (rank : 135) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
IKBL_HUMAN
|
||||||
| NC score | 0.025269 (rank : 136) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.025135 (rank : 137) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.024603 (rank : 138) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.024024 (rank : 139) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
ST18_HUMAN
|
||||||
| NC score | 0.022994 (rank : 140) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O60284 | Gene names | ST18, KIAA0535, ZNF387 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppression of tumorigenicity protein 18 (Zinc finger protein 387). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.022791 (rank : 141) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.022688 (rank : 142) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.022419 (rank : 143) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
MYO1F_HUMAN
|
||||||
| NC score | 0.022384 (rank : 144) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00160, Q8WWN7 | Gene names | MYO1F | |||
|
Domain Architecture |

|
|||||
| Description | Myosin If (Myosin-IE). | |||||
|
MAF_HUMAN
|
||||||
| NC score | 0.020732 (rank : 145) | θ value | 1.81305 (rank : 57) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75444, Q9UP93 | Gene names | MAF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
MAF_MOUSE
|
||||||
| NC score | 0.020315 (rank : 146) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54843 | Gene names | Maf, Maf2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Maf (Proto-oncogene c-maf). | |||||
|
K1H7_HUMAN
|
||||||
| NC score | 0.019531 (rank : 147) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O76014 | Gene names | KRTHA7, HHA7, HKA7 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha7 (Hair keratin, type I Ha7). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.018944 (rank : 148) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.018810 (rank : 149) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
XRN2_HUMAN
|
||||||
| NC score | 0.018509 (rank : 150) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0D6, Q3L8N4, Q6KGZ9, Q9BQL1, Q9NTW0, Q9NXS6, Q9UL53 | Gene names | XRN2 | |||
|
Domain Architecture |

|
|||||
| Description | 5'-3' exoribonuclease 2 (EC 3.1.13.-) (DHM1-like protein) (DHP protein). | |||||
|
MYOC_MOUSE
|
||||||
| NC score | 0.016714 (rank : 151) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O70624, O70289 | Gene names | Myoc, Tigr | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.016035 (rank : 152) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
CAC1H_HUMAN
|
||||||
| NC score | 0.015649 (rank : 153) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95180, O95802, Q8WWI6, Q96QI6, Q96RZ9, Q9NYY4, Q9NYY5 | Gene names | CACNA1H | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2) (Low-voltage-activated calcium channel alpha1 3.2 subunit). | |||||
|
KIF3C_HUMAN
|
||||||
| NC score | 0.014601 (rank : 154) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O14782, O43544 | Gene names | KIF3C | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3C. | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.014072 (rank : 155) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.014013 (rank : 156) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.013780 (rank : 157) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
TRI39_MOUSE
|
||||||
| NC score | 0.013661 (rank : 158) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9ESN2, Q8BPR5, Q8K0F7 | Gene names | Trim39, Rnf23, Tfp | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.012606 (rank : 159) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
TRI39_HUMAN
|
||||||
| NC score | 0.012331 (rank : 160) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9HCM9, Q76BL3, Q8IYT9, Q96IB6 | Gene names | TRIM39, RNF23, TFP | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 39 (RING finger protein 23) (Testis-abundant finger protein). | |||||
|
XKR5_HUMAN
|
||||||
| NC score | 0.011973 (rank : 161) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UX68, Q5GH74 | Gene names | XKR5, XRG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 5. | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.011542 (rank : 162) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.010455 (rank : 163) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
TRI45_MOUSE
|
||||||
| NC score | 0.009484 (rank : 164) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PFY8, Q8BVT5 | Gene names | Trim45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 45. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.009104 (rank : 165) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
DEMA_HUMAN
|
||||||
| NC score | 0.009044 (rank : 166) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08495, Q13215, Q9BRE3 | Gene names | EPB49, DMT | |||
|
Domain Architecture |

|
|||||
| Description | Dematin (Erythrocyte membrane protein band 4.9). | |||||
|
ZMYM3_MOUSE
|
||||||
| NC score | 0.009031 (rank : 167) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.008971 (rank : 168) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.008263 (rank : 169) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
DMPK_HUMAN
|
||||||
| NC score | 0.008256 (rank : 170) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q09013, Q16205 | Gene names | DMPK, MDPK | |||
|
Domain Architecture |

|
|||||
| Description | Myotonin-protein kinase (EC 2.7.11.1) (Myotonic dystrophy protein kinase) (MDPK) (DM-kinase) (DMK) (DMPK) (MT-PK). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.008177 (rank : 171) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
STON1_MOUSE
|
||||||
| NC score | 0.007889 (rank : 172) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
ES8L1_MOUSE
|
||||||
| NC score | 0.007467 (rank : 173) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R5F8, Q8R0D6, Q9D2M6 | Gene names | Eps8l1, Eps8r1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Epidermal growth factor receptor kinase substrate 8-like protein 1 (Epidermal growth factor receptor pathway substrate 8-related protein 1) (EPS8-like protein 1). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.007263 (rank : 174) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.007116 (rank : 175) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PAXI_MOUSE
|
||||||
| NC score | 0.007106 (rank : 176) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI36, Q3TB62, Q3TZQ6, Q8VI37 | Gene names | Pxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paxillin. | |||||
|
ADDB_HUMAN
|
||||||
| NC score | 0.006405 (rank : 177) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P35612, Q13482, Q16412, Q59G82, Q5U5P4, Q6P0P2, Q6PGQ4, Q7Z688, Q7Z689, Q7Z690, Q7Z691 | Gene names | ADD2, ADDB | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.005342 (rank : 178) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
DCJ14_MOUSE
|
||||||
| NC score | 0.005157 (rank : 179) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q921R4, Q3TX73, Q8BUU3, Q9CYB7 | Gene names | Dnajc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DnaJ homolog subfamily C member 14. | |||||
|
LUM_MOUSE
|
||||||
| NC score | 0.005087 (rank : 180) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P51885, Q3TP25, Q99JZ3, Q9CXK0 | Gene names | Lum, Lcn, Ldc | |||
|
Domain Architecture |

|
|||||
| Description | Lumican precursor (Keratan sulfate proteoglycan lumican) (KSPG lumican). | |||||
|
CO8A1_HUMAN
|
||||||
| NC score | 0.004049 (rank : 181) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.003887 (rank : 182) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.003253 (rank : 183) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.001545 (rank : 184) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CEAM8_HUMAN
|
||||||
| NC score | 0.001403 (rank : 185) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 158 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31997, O60399, Q16574 | Gene names | CEACAM8, CGM6 | |||
|
Domain Architecture |

|
|||||
| Description | Carcinoembryonic antigen-related cell adhesion molecule 8 precursor (Carcinoembryonic antigen CGM6) (Nonspecific cross-reacting antigen NCA-95) (CD67 antigen) (CD66b antigen). | |||||