Please be patient as the page loads
|
GIPC2_MOUSE
|
||||||
| SwissProt Accessions | Q9Z2H7, Q9CVG2 | Gene names | Gipc2, Semcap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC2 (SemaF cytoplasmic domain- associated protein 2) (SEMCAP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GIPC2_MOUSE
|
||||||
| θ value | 1.28067e-163 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z2H7, Q9CVG2 | Gene names | Gipc2, Semcap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC2 (SemaF cytoplasmic domain- associated protein 2) (SEMCAP-2). | |||||
|
GIPC2_HUMAN
|
||||||
| θ value | 6.41512e-131 (rank : 2) | NC score | 0.986981 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8TF65, Q8IYD3, Q9NXS7 | Gene names | GIPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC2. | |||||
|
GIPC1_HUMAN
|
||||||
| θ value | 2.15048e-102 (rank : 3) | NC score | 0.972603 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14908, Q9BTC9 | Gene names | GIPC1, C19orf3, GIPC, RGS19IP1 | |||
|
Domain Architecture |
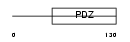
|
|||||
| Description | PDZ domain-containing protein GIPC1 (RGS19-interacting protein 1) (GAIP C-terminus-interacting protein) (RGS-GAIP-interacting protein) (Tax interaction protein 2) (TIP-2). | |||||
|
GIPC1_MOUSE
|
||||||
| θ value | 4.79078e-102 (rank : 4) | NC score | 0.969641 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0G0 | Gene names | Gipc1, Gipc, Rgs19ip1, Semcap1 | |||
|
Domain Architecture |
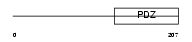
|
|||||
| Description | PDZ domain-containing protein GIPC1 (RGS19-interacting protein 1) (GAIP C-terminus-interacting protein) (RGS-GAIP-interacting protein) (Synectin) (SemaF cytoplasmic domain-associated protein 1) (SEMCAP-1). | |||||
|
GIPC3_HUMAN
|
||||||
| θ value | 2.72669e-89 (rank : 5) | NC score | 0.977810 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TF64, O75227 | Gene names | GIPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC3. | |||||
|
GIPC3_MOUSE
|
||||||
| θ value | 1.49618e-87 (rank : 6) | NC score | 0.976702 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R5M0 | Gene names | Gipc3, Rgs19ip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC3 (Regulator of G-protein signaling 19-interacting protein 3). | |||||
|
SNTA1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 7) | NC score | 0.165366 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13424, Q16438 | Gene names | SNTA1, SNT1 | |||
|
Domain Architecture |
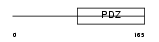
|
|||||
| Description | Alpha-1-syntrophin (59 kDa dystrophin-associated protein A1 acidic component 1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 1) (TACIP1) (Syntrophin 1). | |||||
|
CSKP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.056858 (rank : 129) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
CSKP_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.056814 (rank : 130) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
MAGI2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.174074 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 11) | NC score | 0.175093 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.189213 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
SNTA1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.155280 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61234, Q8VEF3 | Gene names | Snta1, Snt1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-syntrophin (59 kDa dystrophin-associated protein A1 acidic component 1) (Syntrophin 1). | |||||
|
SDCB1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.110700 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |
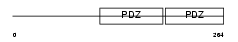
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
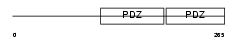
SDCB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.109972 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |
|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
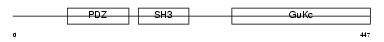
EM55_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.125917 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
GRIP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.181812 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.183027 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.124252 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
CNKR2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.136561 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 21) | NC score | 0.136631 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.170862 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
RHPN1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.083517 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.178109 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
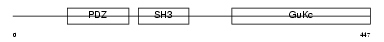
EM55_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.120564 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |
|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.168836 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
PICK1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.145063 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62083 | Gene names | Pick1, Prkcabp | |||
|
Domain Architecture |

|
|||||
| Description | PRKCA-binding protein (Protein kinase C-alpha-binding protein) (Protein interacting with C kinase 1). | |||||
|
PICK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.145936 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NRD5, O95906 | Gene names | PICK1, PRKCABP | |||
|
Domain Architecture |

|
|||||
| Description | PRKCA-binding protein (Protein kinase C-alpha-binding protein) (Protein interacting with C kinase 1). | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.137937 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
GOPC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.138490 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
GOPC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.140797 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BH60, Q8BSV4, Q920R1, Q9ET11 | Gene names | Gopc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.091574 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
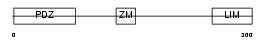
PDLI1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.068568 (rank : 107) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O70400, Q99K93 | Gene names | Pdlim1, Clim1 | |||
|
Domain Architecture |
|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
SNTB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.137395 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99L88, O35925 | Gene names | Sntb1, Snt2b1 | |||
|
Domain Architecture |
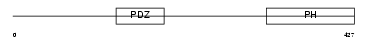
|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Syntrophin 2). | |||||
|
APBA3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.148036 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |
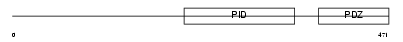
|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
MPP3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.121092 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
MPP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.123318 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88910 | Gene names | Mpp3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Dlgh3 protein). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.129262 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.080644 (rank : 86) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.079417 (rank : 87) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
SNTB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.137741 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13884, O14912, Q4KMG8 | Gene names | SNTB1, SNT2B1 | |||
|
Domain Architecture |
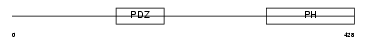
|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Tax interaction protein 43) (TIP-43) (Syntrophin 2) (BSYN2). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 42) | NC score | 0.133294 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.016154 (rank : 138) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.147682 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
LNX1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.128898 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
LNX1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.126046 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70263, O70264, Q8BRI8, Q8CFR3 | Gene names | Lnx1, Lnx | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.150140 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
PDLI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.050407 (rank : 136) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |
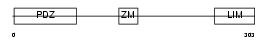
|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.079319 (rank : 88) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.081883 (rank : 82) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.093926 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.145714 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.129861 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
RHPN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.070892 (rank : 103) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
SDCB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.078558 (rank : 89) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99JZ0 | Gene names | Sdcbp2 | |||
|
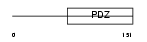
Domain Architecture |
|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.013730 (rank : 139) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.131929 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.081594 (rank : 83) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
SNTG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.119690 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NY99 | Gene names | SNTG2 | |||
|
Domain Architecture |
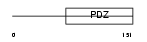
|
|||||
| Description | Gamma-2-syntrophin (G2SYN) (Syntrophin 5) (SYN5). | |||||
|
ARHGC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.062813 (rank : 116) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
DLG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.105961 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
DLG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.105926 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
APBA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.100999 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
DLG4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.097348 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
PDLI4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.043679 (rank : 137) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70271, Q8K0W4 | Gene names | Pdlim4, Ril | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
SNTG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.128609 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NSN8, Q9NY98 | Gene names | SNTG1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-1-syntrophin (G1SYN) (Syntrophin 4) (SYN4). | |||||
|
STAG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.007392 (rank : 140) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.137603 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.067666 (rank : 109) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
SNTG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.127303 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q925E1, Q9D3Z5 | Gene names | Sntg1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-1-syntrophin (G1SYN) (Syntrophin 4) (SYN4). | |||||
|
SNTG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.112173 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q925E0 | Gene names | Sntg2 | |||
|
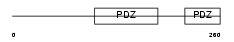
Domain Architecture |
|
|||||
| Description | Gamma-2-syntrophin (G2SYN) (Syntrophin 5) (SYN5). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.075044 (rank : 95) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
APBA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.082077 (rank : 81) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APBA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.082697 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
APXL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.060098 (rank : 120) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ARHGC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.057346 (rank : 125) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
DLG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.087531 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DLG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.087073 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
DLG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.100366 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92796, Q9ULI8 | Gene names | DLG3, KIAA1232 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102) (Neuroendocrine-DLG) (XLMR). | |||||
|
DLG3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.100952 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70175 | Gene names | Dlg3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102). | |||||
|
DLG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.094201 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
GRASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052721 (rank : 135) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
IL16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.099948 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
IL16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.086322 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
K1849_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.070062 (rank : 105) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
K1849_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.068106 (rank : 108) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
LIN7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.110026 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.110003 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
LIN7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.118721 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.116792 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
LIN7C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.121282 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.121282 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LNX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.099273 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N448, Q5W0P0, Q6ZMH2, Q96SH4 | Gene names | LNX2, PDZRN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2) (PDZ domain- containing RING finger protein 1). | |||||
|
LNX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.096219 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91XL2, Q8CBE1 | Gene names | Lnx2 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2). | |||||
|
MPP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.071497 (rank : 101) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
MPP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.072349 (rank : 99) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
MPP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.077224 (rank : 90) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
MPP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.076407 (rank : 91) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
MPP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.080935 (rank : 84) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MPP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.080725 (rank : 85) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
MPP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.073573 (rank : 97) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZW5, Q9H0E1 | Gene names | MPP6, VAM1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Veli-associated MAGUK 1) (VAM-1). | |||||
|
MPP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.075709 (rank : 93) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLB0, Q9JLB1, Q9WV37 | Gene names | Mpp6, Dlgh4, Pals2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Protein associated with Lin-7 2) (Dlgh4 protein) (P55T protein). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.090074 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.088835 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.060615 (rank : 119) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
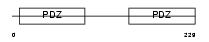
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.067308 (rank : 110) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.065944 (rank : 112) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.111757 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
PAR6A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.054451 (rank : 134) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NPB6, O14911, Q9NPJ7 | Gene names | PARD6A, PAR6A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog alpha (PAR-6 alpha) (PAR-6A) (PAR-6) (PAR6C) (Tax interaction protein 40) (TIP-40). | |||||
|
PAR6A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.054786 (rank : 133) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z101, Q5RL03, Q6P8R2 | Gene names | Pard6a, Par6a | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog alpha (PAR-6 alpha) (PAR-6A) (PAR-6). | |||||
|
PAR6B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057003 (rank : 127) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BYG5, Q9Y510 | Gene names | PARD6B, PAR6B | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PAR6B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.056545 (rank : 131) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JK83, Q8R3J8 | Gene names | Pard6b, Par6b | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PAR6G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.057027 (rank : 126) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BYG4 | Gene names | PARD6G, PAR6G | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog gamma (PAR-6 gamma) (PAR6D). | |||||
|
PAR6G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.057745 (rank : 124) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JK84 | Gene names | Pard6g, Par6g | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog gamma (PAR-6 gamma) (PAR6A). | |||||
|
PDLI2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.059281 (rank : 122) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
PDLI2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.055200 (rank : 132) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1G6 | Gene names | Pdlim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.075296 (rank : 94) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.074484 (rank : 96) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.070607 (rank : 104) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.072461 (rank : 98) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.065153 (rank : 113) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.056858 (rank : 128) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
PDZD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.092806 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.059107 (rank : 123) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.059530 (rank : 121) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.064017 (rank : 114) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
SDCB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.069077 (rank : 106) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H190, O95892, Q5W0X1, Q9BZ42, Q9H567, Q9NRY8 | Gene names | SDCBP2, SITAC18 | |||
|
Domain Architecture |
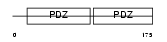
|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
SNTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.110376 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13425, Q9BY09 | Gene names | SNTB2, SNT2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-syntrophin (59 kDa dystrophin-associated protein A1 basic component 2) (Syntrophin 3) (SNT3) (Syntrophin-like) (SNTL). | |||||
|
SNTB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.109550 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61235 | Gene names | Sntb2, Snt2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-syntrophin (59 kDa dystrophin-associated protein A1 basic component 2) (Syntrophin 3) (SNT3) (Syntrophin-like) (SNTL). | |||||
|
SYJ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.097777 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |
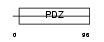
|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
TX1B3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.071622 (rank : 100) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.071363 (rank : 102) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.121311 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.111497 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.060906 (rank : 118) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.062388 (rank : 117) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.076190 (rank : 92) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.082265 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ZO3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.063942 (rank : 115) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ZO3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.066813 (rank : 111) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
GIPC2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.28067e-163 (rank : 1) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9Z2H7, Q9CVG2 | Gene names | Gipc2, Semcap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC2 (SemaF cytoplasmic domain- associated protein 2) (SEMCAP-2). | |||||
|
GIPC2_HUMAN
|
||||||
| NC score | 0.986981 (rank : 2) | θ value | 6.41512e-131 (rank : 2) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8TF65, Q8IYD3, Q9NXS7 | Gene names | GIPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC2. | |||||
|
GIPC3_HUMAN
|
||||||
| NC score | 0.977810 (rank : 3) | θ value | 2.72669e-89 (rank : 5) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TF64, O75227 | Gene names | GIPC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC3. | |||||
|
GIPC3_MOUSE
|
||||||
| NC score | 0.976702 (rank : 4) | θ value | 1.49618e-87 (rank : 6) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R5M0 | Gene names | Gipc3, Rgs19ip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein GIPC3 (Regulator of G-protein signaling 19-interacting protein 3). | |||||
|
GIPC1_HUMAN
|
||||||
| NC score | 0.972603 (rank : 5) | θ value | 2.15048e-102 (rank : 3) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O14908, Q9BTC9 | Gene names | GIPC1, C19orf3, GIPC, RGS19IP1 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ domain-containing protein GIPC1 (RGS19-interacting protein 1) (GAIP C-terminus-interacting protein) (RGS-GAIP-interacting protein) (Tax interaction protein 2) (TIP-2). | |||||
|
GIPC1_MOUSE
|
||||||
| NC score | 0.969641 (rank : 6) | θ value | 4.79078e-102 (rank : 4) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0G0 | Gene names | Gipc1, Gipc, Rgs19ip1, Semcap1 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ domain-containing protein GIPC1 (RGS19-interacting protein 1) (GAIP C-terminus-interacting protein) (RGS-GAIP-interacting protein) (Synectin) (SemaF cytoplasmic domain-associated protein 1) (SEMCAP-1). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.189213 (rank : 7) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.183027 (rank : 8) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP1_HUMAN
|
||||||
| NC score | 0.181812 (rank : 9) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.178109 (rank : 10) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.175093 (rank : 11) | θ value | 0.0431538 (rank : 11) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
MAGI2_HUMAN
|
||||||
| NC score | 0.174074 (rank : 12) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86UL8, O60434, O60510, Q86UI7, Q9NP44, Q9UDQ5, Q9UDU1 | Gene names | MAGI2, ACVRINP1, AIP1, KIAA0705 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Atrophin-1-interacting protein A). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.170862 (rank : 13) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.168836 (rank : 14) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
SNTA1_HUMAN
|
||||||
| NC score | 0.165366 (rank : 15) | θ value | 0.00228821 (rank : 7) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q13424, Q16438 | Gene names | SNTA1, SNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-syntrophin (59 kDa dystrophin-associated protein A1 acidic component 1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 1) (TACIP1) (Syntrophin 1). | |||||
|
SNTA1_MOUSE
|
||||||
| NC score | 0.155280 (rank : 16) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61234, Q8VEF3 | Gene names | Snta1, Snt1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-1-syntrophin (59 kDa dystrophin-associated protein A1 acidic component 1) (Syntrophin 1). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.150140 (rank : 17) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
APBA3_MOUSE
|
||||||
| NC score | 0.148036 (rank : 18) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O88888 | Gene names | Apba3, Mint3 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.147682 (rank : 19) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
PICK1_HUMAN
|
||||||
| NC score | 0.145936 (rank : 20) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9NRD5, O95906 | Gene names | PICK1, PRKCABP | |||
|
Domain Architecture |

|
|||||
| Description | PRKCA-binding protein (Protein kinase C-alpha-binding protein) (Protein interacting with C kinase 1). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.145714 (rank : 21) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PICK1_MOUSE
|
||||||
| NC score | 0.145063 (rank : 22) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q62083 | Gene names | Pick1, Prkcabp | |||
|
Domain Architecture |

|
|||||
| Description | PRKCA-binding protein (Protein kinase C-alpha-binding protein) (Protein interacting with C kinase 1). | |||||
|
GOPC_MOUSE
|
||||||
| NC score | 0.140797 (rank : 23) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8BH60, Q8BSV4, Q920R1, Q9ET11 | Gene names | Gopc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST). | |||||
|
GOPC_HUMAN
|
||||||
| NC score | 0.138490 (rank : 24) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HD26, Q59FS4, Q969U8 | Gene names | GOPC, CAL, FIG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi-associated PDZ and coiled-coil motif-containing protein (PDZ protein interacting specifically with TC10) (PIST) (CFTR-associated ligand) (Fused in glioblastoma). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.137937 (rank : 25) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
SNTB1_HUMAN
|
||||||
| NC score | 0.137741 (rank : 26) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q13884, O14912, Q4KMG8 | Gene names | SNTB1, SNT2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Tax interaction protein 43) (TIP-43) (Syntrophin 2) (BSYN2). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.137603 (rank : 27) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
SNTB1_MOUSE
|
||||||
| NC score | 0.137395 (rank : 28) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q99L88, O35925 | Gene names | Sntb1, Snt2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1-syntrophin (59 kDa dystrophin-associated protein A1 basic component 1) (DAPA1B) (Syntrophin 2). | |||||
|
CNKR2_MOUSE
|
||||||
| NC score | 0.136631 (rank : 29) | θ value | 0.279714 (rank : 21) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q80YA9, Q80TP2 | Gene names | Cnksr2, Kiaa0902 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
CNKR2_HUMAN
|
||||||
| NC score | 0.136561 (rank : 30) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WXI2, O94976, Q8WXI1 | Gene names | CNKSR2, CNK2, KIAA0902, KSR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Connector enhancer of kinase suppressor of ras 2 (Connector enhancer of KSR2) (CNK2). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.133294 (rank : 31) | θ value | 1.06291 (rank : 42) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.131929 (rank : 32) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.129861 (rank : 33) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.129262 (rank : 34) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
LNX1_HUMAN
|
||||||
| NC score | 0.128898 (rank : 35) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
SNTG1_HUMAN
|
||||||
| NC score | 0.128609 (rank : 36) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NSN8, Q9NY98 | Gene names | SNTG1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-1-syntrophin (G1SYN) (Syntrophin 4) (SYN4). | |||||
|
SNTG1_MOUSE
|
||||||
| NC score | 0.127303 (rank : 37) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q925E1, Q9D3Z5 | Gene names | Sntg1 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-1-syntrophin (G1SYN) (Syntrophin 4) (SYN4). | |||||
|
LNX1_MOUSE
|
||||||
| NC score | 0.126046 (rank : 38) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O70263, O70264, Q8BRI8, Q8CFR3 | Gene names | Lnx1, Lnx | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
EM55_MOUSE
|
||||||
| NC score | 0.125917 (rank : 39) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70290 | Gene names | Mpp1 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1) (Palmitoylated protein p55). | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.124252 (rank : 40) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
MPP3_MOUSE
|
||||||
| NC score | 0.123318 (rank : 41) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O88910 | Gene names | Mpp3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Dlgh3 protein). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.121311 (rank : 42) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
LIN7C_HUMAN
|
||||||
| NC score | 0.121282 (rank : 43) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| NC score | 0.121282 (rank : 44) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
MPP3_HUMAN
|
||||||
| NC score | 0.121092 (rank : 45) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13368 | Gene names | MPP3, DLG3 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 3 (Protein MPP3) (Discs large homolog 3). | |||||
|
EM55_HUMAN
|
||||||
| NC score | 0.120564 (rank : 46) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q00013 | Gene names | MPP1, DXS552E, EMP55 | |||
|
Domain Architecture |

|
|||||
| Description | 55 kDa erythrocyte membrane protein (p55) (Membrane protein, palmitoylated 1). | |||||
|
SNTG2_HUMAN
|
||||||
| NC score | 0.119690 (rank : 47) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9NY99 | Gene names | SNTG2 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-2-syntrophin (G2SYN) (Syntrophin 5) (SYN5). | |||||
|
LIN7B_HUMAN
|
||||||
| NC score | 0.118721 (rank : 48) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7B_MOUSE
|
||||||
| NC score | 0.116792 (rank : 49) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
SNTG2_MOUSE
|
||||||
| NC score | 0.112173 (rank : 50) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q925E0 | Gene names | Sntg2 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-2-syntrophin (G2SYN) (Syntrophin 5) (SYN5). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.111757 (rank : 51) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.111497 (rank : 52) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
SDCB1_HUMAN
|
||||||
| NC score | 0.110700 (rank : 53) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00560, O00173, O43391 | Gene names | SDCBP, MDA9, SYCL | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Melanoma differentiation- associated protein 9) (MDA-9) (Scaffold protein Pbp1) (Pro-TGF-alpha cytoplasmic domain-interacting protein 18) (TACIP18). | |||||
|
SNTB2_HUMAN
|
||||||
| NC score | 0.110376 (rank : 54) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13425, Q9BY09 | Gene names | SNTB2, SNT2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-syntrophin (59 kDa dystrophin-associated protein A1 basic component 2) (Syntrophin 3) (SNT3) (Syntrophin-like) (SNTL). | |||||
|
LIN7A_HUMAN
|
||||||
| NC score | 0.110026 (rank : 55) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| NC score | 0.110003 (rank : 56) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
SDCB1_MOUSE
|
||||||
| NC score | 0.109972 (rank : 57) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O08992, Q544P5 | Gene names | Sdcbp | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-1 (Syndecan-binding protein 1) (Scaffold protein Pbp1). | |||||
|
SNTB2_MOUSE
|
||||||
| NC score | 0.109550 (rank : 58) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61235 | Gene names | Sntb2, Snt2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-2-syntrophin (59 kDa dystrophin-associated protein A1 basic component 2) (Syntrophin 3) (SNT3) (Syntrophin-like) (SNTL). | |||||
|
DLG2_HUMAN
|
||||||
| NC score | 0.105961 (rank : 59) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
DLG2_MOUSE
|
||||||
| NC score | 0.105926 (rank : 60) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91XM9, Q8BXK7, Q8BYG5 | Gene names | Dlg2, Dlgh2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
APBA1_HUMAN
|
||||||
| NC score | 0.100999 (rank : 61) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q02410, O14914, O60570, Q5VYR8 | Gene names | APBA1, MINT1, X11 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 1 (Neuron- specific X11 protein) (Neuronal Munc18-1-interacting protein 1) (Mint- 1) (Adapter protein X11alpha). | |||||
|
DLG3_MOUSE
|
||||||
| NC score | 0.100952 (rank : 62) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P70175 | Gene names | Dlg3, Dlgh3 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102). | |||||
|
DLG3_HUMAN
|
||||||
| NC score | 0.100366 (rank : 63) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q92796, Q9ULI8 | Gene names | DLG3, KIAA1232 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 3 (Synapse-associated protein 102) (SAP102) (Neuroendocrine-DLG) (XLMR). | |||||
|
IL16_HUMAN
|
||||||
| NC score | 0.099948 (rank : 64) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q14005, Q16435, Q9UP18 | Gene names | IL16 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
LNX2_HUMAN
|
||||||
| NC score | 0.099273 (rank : 65) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8N448, Q5W0P0, Q6ZMH2, Q96SH4 | Gene names | LNX2, PDZRN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2) (PDZ domain- containing RING finger protein 1). | |||||
|
SYJ2B_HUMAN
|
||||||
| NC score | 0.097777 (rank : 66) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P57105, Q96IA4 | Gene names | SYNJ2BP, OMP25 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-2-binding protein (Mitochondrial outer membrane protein 25). | |||||
|
DLG4_MOUSE
|
||||||
| NC score | 0.097348 (rank : 67) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q62108, Q5NCV5, Q5NCV6, Q5NCV7, Q91WJ1 | Gene names | Dlg4, Dlgh4, Psd95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
LNX2_MOUSE
|
||||||
| NC score | 0.096219 (rank : 68) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91XL2, Q8CBE1 | Gene names | Lnx2 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2). | |||||
|
DLG4_HUMAN
|
||||||
| NC score | 0.094201 (rank : 69) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P78352, Q92941, Q9UKK8 | Gene names | DLG4, PSD95 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 4 (Postsynaptic density protein 95) (PSD-95) (Synapse-associated protein 90) (SAP90). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.093926 (rank : 70) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
PDZD7_HUMAN
|
||||||
| NC score | 0.092806 (rank : 71) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.091574 (rank : 72) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.090074 (rank : 73) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.088835 (rank : 74) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
DLG1_HUMAN
|
||||||
| NC score | 0.087531 (rank : 75) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q12959, Q12958 | Gene names | DLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (hDlg). | |||||
|
DLG1_MOUSE
|
||||||
| NC score | 0.087073 (rank : 76) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q811D0, Q62402, Q6PGB5, Q8CGN7 | Gene names | Dlg1, Dlgh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large homolog 1 (Synapse-associated protein 97) (SAP-97) (Embryo-dlg/synapse-associated protein 97) (E-dlg/SAP97). | |||||
|
IL16_MOUSE
|
||||||
| NC score | 0.086322 (rank : 77) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O54824, O70236, Q3TEM4, Q8R0G5 | Gene names | Il16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-16 precursor (IL-16) (Lymphocyte chemoattractant factor) (LCF). | |||||
|
RHPN1_MOUSE
|
||||||
| NC score | 0.083517 (rank : 78) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61085 | Gene names | Rhpn1, Grbp | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
APBA2_MOUSE
|
||||||
| NC score | 0.082697 (rank : 79) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P98084, Q6PAJ2 | Gene names | Apba2, X11l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.082265 (rank : 80) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
APBA2_HUMAN
|
||||||
| NC score | 0.082077 (rank : 81) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q99767, O60571 | Gene names | APBA2, MINT2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 2 (Neuron- specific X11L protein) (Neuronal Munc18-1-interacting protein 2) (Mint-2) (Adapter protein X11beta). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.081883 (rank : 82) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.081594 (rank : 83) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
MPP5_HUMAN
|
||||||
| NC score | 0.080935 (rank : 84) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
MPP5_MOUSE
|
||||||
| NC score | 0.080725 (rank : 85) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.080644 (rank : 86) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.079417 (rank : 87) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.079319 (rank : 88) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
SDCB2_MOUSE
|
||||||
| NC score | 0.078558 (rank : 89) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99JZ0 | Gene names | Sdcbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
MPP4_HUMAN
|
||||||
| NC score | 0.077224 (rank : 90) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96JB8, Q6ZNH6, Q96Q43, Q96Q44 | Gene names | MPP4, ALS2CR5, DLG6 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 5 protein). | |||||
|
MPP4_MOUSE
|
||||||
| NC score | 0.076407 (rank : 91) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q6P7F1, Q8BTT3, Q8BXM5, Q920P7, Q920P8 | Gene names | Mpp4, Dlg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAGUK p55 subfamily member 4 (Discs large homolog 6) (MDLG6). | |||||
|
ZO2_HUMAN
|
||||||
| NC score | 0.076190 (rank : 92) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9UDY2, Q15883, Q8N756, Q8NI14, Q99839, Q9UDY0, Q9UDY1 | Gene names | TJP2, X104, ZO2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
MPP6_MOUSE
|
||||||
| NC score | 0.075709 (rank : 93) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9JLB0, Q9JLB1, Q9WV37 | Gene names | Mpp6, Dlgh4, Pals2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Protein associated with Lin-7 2) (Dlgh4 protein) (P55T protein). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.075296 (rank : 94) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.075044 (rank : 95) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.074484 (rank : 96) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
MPP6_HUMAN
|
||||||
| NC score | 0.073573 (rank : 97) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZW5, Q9H0E1 | Gene names | MPP6, VAM1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 6 (Veli-associated MAGUK 1) (VAM-1). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.072461 (rank : 98) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
MPP2_MOUSE
|
||||||
| NC score | 0.072349 (rank : 99) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9WV34 | Gene names | Mpp2, Dlgh2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Dlgh2 protein). | |||||
|
TX1B3_HUMAN
|
||||||
| NC score | 0.071622 (rank : 100) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
MPP2_HUMAN
|
||||||
| NC score | 0.071497 (rank : 101) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14168, Q9BQJ2 | Gene names | MPP2, DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 2 (Protein MPP2) (Discs large homolog 2). | |||||
|
TX1B3_MOUSE
|
||||||
| NC score | 0.071363 (rank : 102) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
RHPN1_HUMAN
|
||||||
| NC score | 0.070892 (rank : 103) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TCX5, Q8TAV1 | Gene names | RHPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhophilin-1 (GTP-Rho-binding protein 1). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.070607 (rank : 104) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
K1849_HUMAN
|
||||||
| NC score | 0.070062 (rank : 105) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96JH8, Q75LH3, Q9BSP5, Q9H0M6, Q9NW43, Q9NWC4 | Gene names | KIAA1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
SDCB2_HUMAN
|
||||||
| NC score | 0.069077 (rank : 106) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H190, O95892, Q5W0X1, Q9BZ42, Q9H567, Q9NRY8 | Gene names | SDCBP2, SITAC18 | |||
|
Domain Architecture |

|
|||||
| Description | Syntenin-2 (Syndecan-binding protein 2). | |||||
|
PDLI1_MOUSE
|
||||||
| NC score | 0.068568 (rank : 107) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O70400, Q99K93 | Gene names | Pdlim1, Clim1 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.068106 (rank : 108) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.067666 (rank : 109) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.067308 (rank : 110) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
ZO3_MOUSE
|
||||||
| NC score | 0.066813 (rank : 111) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QXY1 | Gene names | Tjp3, Zo3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.065944 (rank : 112) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.065153 (rank : 113) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.064017 (rank : 114) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
ZO3_HUMAN
|
||||||
| NC score | 0.063942 (rank : 115) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95049 | Gene names | TJP3, ZO3 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-3 (Zonula occludens 3 protein) (Zona occludens 3 protein) (Tight junction protein 3). | |||||
|
ARHGC_HUMAN
|
||||||
| NC score | 0.062813 (rank : 116) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NZN5, O15086 | Gene names | ARHGEF12, KIAA0382, LARG | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
ZO1_MOUSE
|
||||||
| NC score | 0.062388 (rank : 117) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P39447 | Gene names | Tjp1, Zo1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.060906 (rank : 118) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.060615 (rank : 119) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.060098 (rank : 120) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.059530 (rank : 121) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PDLI2_HUMAN
|
||||||
| NC score | 0.059281 (rank : 122) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96JY6, Q7Z584, Q86WM8, Q8WZ29, Q9H4L9, Q9H7I2 | Gene names | PDLIM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique) (PDZ-LIM protein). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.059107 (rank : 123) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PAR6G_MOUSE
|
||||||
| NC score | 0.057745 (rank : 124) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9JK84 | Gene names | Pard6g, Par6g | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog gamma (PAR-6 gamma) (PAR6A). | |||||
|
ARHGC_MOUSE
|
||||||
| NC score | 0.057346 (rank : 125) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8R4H2, Q80U18 | Gene names | Arhgef12, Kiaa0382, Larg | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 12 (Leukemia-associated RhoGEF). | |||||
|
PAR6G_HUMAN
|
||||||
| NC score | 0.057027 (rank : 126) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9BYG4 | Gene names | PARD6G, PAR6G | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog gamma (PAR-6 gamma) (PAR6D). | |||||
|
PAR6B_HUMAN
|
||||||
| NC score | 0.057003 (rank : 127) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BYG5, Q9Y510 | Gene names | PARD6B, PAR6B | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.056858 (rank : 128) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
CSKP_HUMAN
|
||||||
| NC score | 0.056858 (rank : 129) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1022 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14936, O43215, Q9NYB3 | Gene names | CASK, LIN2 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (hCASK) (Calcium/calmodulin-dependent serine protein kinase) (Lin-2 homolog). | |||||
|
CSKP_MOUSE
|
||||||
| NC score | 0.056814 (rank : 130) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O70589, O70588 | Gene names | Cask | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral plasma membrane protein CASK (EC 2.7.11.1) (Calcium/calmodulin-dependent serine protein kinase). | |||||
|
PAR6B_MOUSE
|
||||||
| NC score | 0.056545 (rank : 131) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9JK83, Q8R3J8 | Gene names | Pard6b, Par6b | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog beta (PAR-6 beta) (PAR-6B). | |||||
|
PDLI2_MOUSE
|
||||||
| NC score | 0.055200 (rank : 132) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1G6 | Gene names | Pdlim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 2 (PDZ-LIM protein mystique). | |||||
|
PAR6A_MOUSE
|
||||||
| NC score | 0.054786 (rank : 133) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9Z101, Q5RL03, Q6P8R2 | Gene names | Pard6a, Par6a | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog alpha (PAR-6 alpha) (PAR-6A) (PAR-6). | |||||
|
PAR6A_HUMAN
|
||||||
| NC score | 0.054451 (rank : 134) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9NPB6, O14911, Q9NPJ7 | Gene names | PARD6A, PAR6A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning defective 6 homolog alpha (PAR-6 alpha) (PAR-6A) (PAR-6) (PAR6C) (Tax interaction protein 40) (TIP-40). | |||||
|
GRASP_HUMAN
|
||||||
| NC score | 0.052721 (rank : 135) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
PDLI1_HUMAN
|
||||||
| NC score | 0.050407 (rank : 136) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
PDLI4_MOUSE
|
||||||
| NC score | 0.043679 (rank : 137) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P70271, Q8K0W4 | Gene names | Pdlim4, Ril | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.016154 (rank : 138) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.013730 (rank : 139) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
STAG1_MOUSE
|
||||||
| NC score | 0.007392 (rank : 140) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 71 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D3E6, O08982 | Gene names | Stag1, Sa1 | |||
|
Domain Architecture |

|
|||||
| Description | Cohesin subunit SA-1 (Stromal antigen 1) (SCC3 homolog 1). | |||||