Please be patient as the page loads
|
P3C2B_HUMAN
|
||||||
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P3C2A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979901 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.980410 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.887741 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.901667 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 1.1019e-106 (rank : 6) | NC score | 0.811608 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_HUMAN
|
||||||
| θ value | 1.43912e-106 (rank : 7) | NC score | 0.811498 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 9.65309e-103 (rank : 8) | NC score | 0.811600 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | 2.01279e-100 (rank : 9) | NC score | 0.812177 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 7.91507e-97 (rank : 10) | NC score | 0.801978 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 6.7005e-96 (rank : 11) | NC score | 0.801474 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 6.93392e-93 (rank : 12) | NC score | 0.798980 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 7.66637e-92 (rank : 13) | NC score | 0.798749 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 14) | NC score | 0.734868 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 9.83387e-39 (rank : 15) | NC score | 0.733676 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 16) | NC score | 0.709246 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 17) | NC score | 0.666821 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 2.35151e-24 (rank : 18) | NC score | 0.667948 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 19) | NC score | 0.477898 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 20) | NC score | 0.490903 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 21) | NC score | 0.482855 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SYT7_MOUSE
|
||||||
| θ value | 1.99529e-15 (rank : 22) | NC score | 0.491055 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |
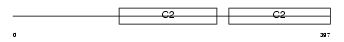
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
DOC2B_HUMAN
|
||||||
| θ value | 2.60593e-15 (rank : 23) | NC score | 0.501817 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 24) | NC score | 0.466159 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
DOC2B_MOUSE
|
||||||
| θ value | 5.8054e-15 (rank : 25) | NC score | 0.499783 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2A_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 26) | NC score | 0.499707 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 27) | NC score | 0.504834 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
DOC2A_HUMAN
|
||||||
| θ value | 1.42992e-13 (rank : 28) | NC score | 0.499957 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 29) | NC score | 0.509347 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | 5.43371e-13 (rank : 30) | NC score | 0.449884 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | 7.09661e-13 (rank : 31) | NC score | 0.503658 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 32) | NC score | 0.343959 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 33) | NC score | 0.436756 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | 2.69671e-12 (rank : 34) | NC score | 0.440344 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
SYT2_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 35) | NC score | 0.473809 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |
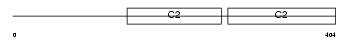
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 36) | NC score | 0.470609 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |
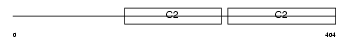
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 37) | NC score | 0.470599 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |
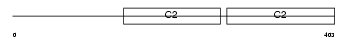
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT2_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 38) | NC score | 0.473778 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 39) | NC score | 0.511221 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL4_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 40) | NC score | 0.507899 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 41) | NC score | 0.323558 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 42) | NC score | 0.488090 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
RIMS4_MOUSE
|
||||||
| θ value | 2.98157e-11 (rank : 43) | NC score | 0.449171 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
SYT5_HUMAN
|
||||||
| θ value | 2.98157e-11 (rank : 44) | NC score | 0.472588 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
RIMS4_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 45) | NC score | 0.447776 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
SYT7_HUMAN
|
||||||
| θ value | 6.64225e-11 (rank : 46) | NC score | 0.476984 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 47) | NC score | 0.467543 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT6_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 48) | NC score | 0.466489 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT6_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 49) | NC score | 0.464695 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
KPCG_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 50) | NC score | 0.135202 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
SYT3_MOUSE
|
||||||
| θ value | 7.34386e-10 (rank : 51) | NC score | 0.465971 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT4_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 52) | NC score | 0.463927 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
KPCA_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 53) | NC score | 0.128977 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
SYT11_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 54) | NC score | 0.462802 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 55) | NC score | 0.463335 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT9_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 56) | NC score | 0.457142 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 57) | NC score | 0.514011 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
KPCG_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 58) | NC score | 0.134052 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
SYT10_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 59) | NC score | 0.458046 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT12_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 60) | NC score | 0.468822 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT4_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 61) | NC score | 0.461500 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
KPCA_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 62) | NC score | 0.125486 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
SYT12_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 63) | NC score | 0.467045 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT10_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 64) | NC score | 0.457612 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT9_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 65) | NC score | 0.455174 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
KPCB_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 66) | NC score | 0.124061 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCB_MOUSE
|
||||||
| θ value | 1.99992e-07 (rank : 67) | NC score | 0.124037 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
RIMS3_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 68) | NC score | 0.380782 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
SYT5_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 69) | NC score | 0.463769 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
RIMS3_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 70) | NC score | 0.374874 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 71) | NC score | 0.479542 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 72) | NC score | 0.483614 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 73) | NC score | 0.326164 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 74) | NC score | 0.327066 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
DOC2G_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 75) | NC score | 0.477394 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
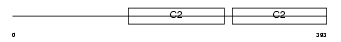
SYT15_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 76) | NC score | 0.462011 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 77) | NC score | 0.164436 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
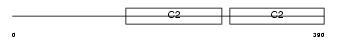
SYT15_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 78) | NC score | 0.433553 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
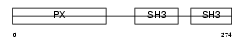
NCF1_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 79) | NC score | 0.101927 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q09014, O70144, Q9JI34 | Gene names | Ncf1 | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil cytosol factor 1 (NCF-1) (Neutrophil NADPH oxidase factor 1) (47 kDa neutrophil oxidase factor) (p47-phox) (NCF-47K). | |||||
|
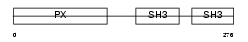
NCF1_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 80) | NC score | 0.109526 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14598, O43842, Q2PP07, Q9BU90, Q9BXI7, Q9BXI8, Q9UDV9, Q9UMU2 | Gene names | NCF1 | |||
|
Domain Architecture |
|
|||||
| Description | Neutrophil cytosol factor 1 (NCF-1) (Neutrophil NADPH oxidase factor 1) (47 kDa neutrophil oxidase factor) (p47-phox) (NCF-47K) (47 kDa autosomal chronic granulomatous disease protein) (NOXO2). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 81) | NC score | 0.294297 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
SNX18_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 82) | NC score | 0.070351 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96RF0 | Gene names | SNAG1, SNX18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
SNX18_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 83) | NC score | 0.066900 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZR2 | Gene names | Snag1, Snx18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 84) | NC score | 0.232470 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RASL2_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 85) | NC score | 0.314357 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 86) | NC score | 0.223684 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 87) | NC score | 0.285459 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
SYT8_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 88) | NC score | 0.427121 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 89) | NC score | 0.227566 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
K0528_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 90) | NC score | 0.258516 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 91) | NC score | 0.258404 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 92) | NC score | 0.209984 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 93) | NC score | 0.211755 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
NCF4_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 94) | NC score | 0.052410 (rank : 169) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97369 | Gene names | Ncf4 | |||
|
Domain Architecture |
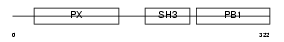
|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 95) | NC score | 0.163516 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
SNX22_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 96) | NC score | 0.063674 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96L94, Q9H844 | Gene names | SNX22 | |||
|
Domain Architecture |
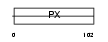
|
|||||
| Description | Sorting nexin-22. | |||||
|
ATR_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 97) | NC score | 0.255631 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
SNX16_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 98) | NC score | 0.088090 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C080 | Gene names | Snx16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-16. | |||||
|
ATR_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 99) | NC score | 0.255712 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 100) | NC score | 0.153763 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
DYSF_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 101) | NC score | 0.197320 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
NCF4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 102) | NC score | 0.063844 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15080, O60808, Q86U56, Q9BU98, Q9NP45 | Gene names | NCF4 | |||
|
Domain Architecture |
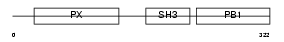
|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 103) | NC score | 0.191720 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
SGK3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 104) | NC score | 0.027701 (rank : 172) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ERE3, Q3UAY2 | Gene names | Sgk3, Cisk, Sgkl | |||
|
Domain Architecture |
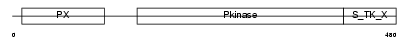
|
|||||
| Description | Serine/threonine-protein kinase Sgk3 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 3) (Serum/glucocorticoid- regulated kinase-like) (Cytokine-independent survival kinase). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 105) | NC score | 0.013569 (rank : 178) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
SGK3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 106) | NC score | 0.027630 (rank : 174) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96BR1, Q9P1Q7, Q9UKG5 | Gene names | SGK3, SGKL | |||
|
Domain Architecture |
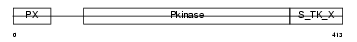
|
|||||
| Description | Serine/threonine-protein kinase Sgk3 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 3) (Serum/glucocorticoid- regulated kinase-like). | |||||
|
SYT13_MOUSE
|
||||||
| θ value | 0.125558 (rank : 107) | NC score | 0.341954 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 0.163984 (rank : 108) | NC score | 0.176693 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 0.163984 (rank : 109) | NC score | 0.173539 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
RASA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 110) | NC score | 0.043704 (rank : 171) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
SNX16_HUMAN
|
||||||
| θ value | 0.279714 (rank : 111) | NC score | 0.085121 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57768, Q8N4U3 | Gene names | SNX16 | |||
|
Domain Architecture |
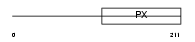
|
|||||
| Description | Sorting nexin-16. | |||||
|
SYT14_HUMAN
|
||||||
| θ value | 0.62314 (rank : 112) | NC score | 0.327107 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.813845 (rank : 113) | NC score | 0.019297 (rank : 176) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
KKCC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 114) | NC score | 0.010120 (rank : 186) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C078, Q80TS0, Q8BXM8, Q8C0G3, Q8CH42, Q8QZT7 | Gene names | Camkk2, Kiaa0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
NOXO1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 115) | NC score | 0.054649 (rank : 166) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VCM2, Q3TZA4, Q8BH41, Q9D747 | Gene names | Noxo1, Snx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase organizer 1 (Nox organizer 1) (Sorting nexin-28). | |||||
|
SNX24_HUMAN
|
||||||
| θ value | 0.813845 (rank : 116) | NC score | 0.054762 (rank : 165) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y343, Q6UY33 | Gene names | SNX24 | |||
|
Domain Architecture |
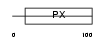
|
|||||
| Description | Sorting nexin-24. | |||||
|
SNX24_MOUSE
|
||||||
| θ value | 0.813845 (rank : 117) | NC score | 0.054170 (rank : 167) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CRB0 | Gene names | Snx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-24. | |||||
|
ATM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 118) | NC score | 0.232768 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 119) | NC score | 0.014690 (rank : 177) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 120) | NC score | 0.161566 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
SYT14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 121) | NC score | 0.326825 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 122) | NC score | 0.013031 (rank : 180) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
HHIP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 123) | NC score | 0.011834 (rank : 182) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TN16, Q8C0B0, Q9WU59 | Gene names | Hhip, Hip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 124) | NC score | 0.002866 (rank : 190) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 125) | NC score | 0.011424 (rank : 184) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
HHIP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 126) | NC score | 0.010481 (rank : 185) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
IQEC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 127) | NC score | 0.011821 (rank : 183) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
IQEC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 128) | NC score | 0.012171 (rank : 181) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
OTOF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.151831 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
WWC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.066312 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
ABCAD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 131) | NC score | 0.001816 (rank : 192) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
ATM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 132) | NC score | 0.223266 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
OTOF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 133) | NC score | 0.153717 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 5.27518 (rank : 134) | NC score | 0.000739 (rank : 193) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ABCG4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.002345 (rank : 191) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H172 | Gene names | ABCG4, WHITE2 | |||
|
Domain Architecture |
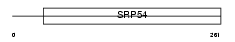
|
|||||
| Description | ATP-binding cassette sub-family G member 4. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | -0.003384 (rank : 196) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
LPIN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.004210 (rank : 189) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14693 | Gene names | LPIN1, KIAA0188 | |||
|
Domain Architecture |
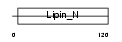
|
|||||
| Description | Lipin-1. | |||||
|
OPSG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | -0.004917 (rank : 197) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35599, Q548Z4 | Gene names | Opn1mw, Gcp | |||
|
Domain Architecture |
|
|||||
| Description | Green-sensitive opsin (Green cone photoreceptor pigment) (Medium wavelength-sensitive cone opsin) (M opsin). | |||||
|
SNX14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.026517 (rank : 175) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 140) | NC score | 0.000273 (rank : 194) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
AIOL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | -0.006912 (rank : 198) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKT9, Q8WWQ9, Q8WWR0, Q8WWR1, Q8WWR2, Q8WWR3 | Gene names | IKZF3, ZNFN1A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
CAC1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.000217 (rank : 195) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.008967 (rank : 188) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.013230 (rank : 179) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.010008 (rank : 187) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PA24A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.094229 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P47712 | Gene names | PLA2G4A, CPLA2, PLA2G4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
SNX14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.027665 (rank : 173) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
UN13B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.166558 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
BAIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.229865 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
BAIP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.227986 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
CPNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.070196 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99829, Q6IBL3, Q9H243, Q9NTZ7 | Gene names | CPNE1, CPN1 | |||
|
Domain Architecture |
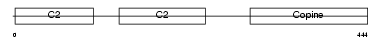
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.084898 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |
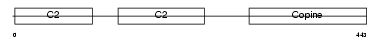
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.084474 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96FN4, Q68D19, Q86XP9 | Gene names | CPNE2 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.084495 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P59108, Q8K1D7 | Gene names | Cpne2 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.097106 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75131, Q8IYA1 | Gene names | CPNE3, CPN3, KIAA0636 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.097265 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BT60, Q5CZX9, Q5DU22 | Gene names | Cpne3, Kiaa0636 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.052457 (rank : 168) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.051008 (rank : 170) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
CPNE5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.088477 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.088752 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.062733 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95741, Q53HA6, Q8WVG1 | Gene names | CPNE6 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
CPNE6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.059058 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z140 | Gene names | Cpne6 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
CPNE7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.075413 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
CPNE8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.082802 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
CPNE8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.080832 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.057959 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.058354 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.083119 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.074939 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
MELPH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.069092 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.070699 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
MYOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.179029 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.076976 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.080903 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
PA24A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.099865 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
PA24B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.091774 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PA24B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.117004 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PA24D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.135471 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
PA24D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.160090 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q50L43, Q3TJC5 | Gene names | Pla2g4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
PA24E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.094751 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3MJ16, Q6ZSC0 | Gene names | PLA2G4E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
PA24E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.107512 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q50L42, Q8BX44 | Gene names | Pla2g4e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
PA24F_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.056152 (rank : 164) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q68DD2, Q6ZMC8 | Gene names | PLA2G4F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
PA24F_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.070860 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q50L41, Q80VQ8, Q80VV9 | Gene names | Pla2g4f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
PLCD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.071500 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.166787 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RASL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.245182 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.237910 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RFIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.057415 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.062919 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.154254 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.059421 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.058453 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.058413 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SYT13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.335361 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
TAC2N_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.171345 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N9U0 | Gene names | MTAC2D1, C14orf47 | |||
|
Domain Architecture |
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
TAC2N_MOUSE
|
||||||
| θ value | θ > 10 (rank : 196) | NC score | 0.170709 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 197) | NC score | 0.168270 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 198) | NC score | 0.239687 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.980410 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.979901 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.901667 (rank : 4) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.887741 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.812177 (rank : 6) | θ value | 2.01279e-100 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.811608 (rank : 7) | θ value | 1.1019e-106 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.811600 (rank : 8) | θ value | 9.65309e-103 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.811498 (rank : 9) | θ value | 1.43912e-106 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.801978 (rank : 10) | θ value | 7.91507e-97 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.801474 (rank : 11) | θ value | 6.7005e-96 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.798980 (rank : 12) | θ value | 6.93392e-93 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.798749 (rank : 13) | θ value | 7.66637e-92 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.734868 (rank : 14) | θ value | 5.76516e-39 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.733676 (rank : 15) | θ value | 9.83387e-39 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.709246 (rank : 16) | θ value | 2.86786e-30 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.667948 (rank : 17) | θ value | 2.35151e-24 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.666821 (rank : 18) | θ value | 6.18819e-25 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.514011 (rank : 19) | θ value | 9.59137e-10 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.511221 (rank : 20) | θ value | 3.52202e-12 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.509347 (rank : 21) | θ value | 2.43908e-13 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL4_MOUSE
|
||||||
| NC score | 0.507899 (rank : 22) | θ value | 7.84624e-12 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.504834 (rank : 23) | θ value | 8.38298e-14 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.503658 (rank : 24) | θ value | 7.09661e-13 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
DOC2B_HUMAN
|
||||||
| NC score | 0.501817 (rank : 25) | θ value | 2.60593e-15 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.499957 (rank : 26) | θ value | 1.42992e-13 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2B_MOUSE
|
||||||
| NC score | 0.499783 (rank : 27) | θ value | 5.8054e-15 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2A_MOUSE
|
||||||
| NC score | 0.499707 (rank : 28) | θ value | 4.91457e-14 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
SYT7_MOUSE
|
||||||
| NC score | 0.491055 (rank : 29) | θ value | 1.99529e-15 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.490903 (rank : 30) | θ value | 8.95645e-16 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.488090 (rank : 31) | θ value | 1.33837e-11 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.483614 (rank : 32) | θ value | 1.29631e-06 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.482855 (rank : 33) | θ value | 8.95645e-16 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.479542 (rank : 34) | θ value | 9.92553e-07 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.477898 (rank : 35) | θ value | 1.058e-16 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
DOC2G_MOUSE
|
||||||
| NC score | 0.477394 (rank : 36) | θ value | 6.43352e-06 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
SYT7_HUMAN
|
||||||
| NC score | 0.476984 (rank : 37) | θ value | 6.64225e-11 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT2_MOUSE
|
||||||
| NC score | 0.473809 (rank : 38) | θ value | 2.69671e-12 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_HUMAN
|
||||||
| NC score | 0.473778 (rank : 39) | θ value | 3.52202e-12 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT5_HUMAN
|
||||||
| NC score | 0.472588 (rank : 40) | θ value | 2.98157e-11 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT1_HUMAN
|
||||||
| NC score | 0.470609 (rank : 41) | θ value | 3.52202e-12 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| NC score | 0.470599 (rank : 42) | θ value | 3.52202e-12 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT12_HUMAN
|
||||||
| NC score | 0.468822 (rank : 43) | θ value | 3.64472e-09 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.467543 (rank : 44) | θ value | 1.9326e-10 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT12_MOUSE
|
||||||
| NC score | 0.467045 (rank : 45) | θ value | 8.11959e-09 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT6_MOUSE
|
||||||
| NC score | 0.466489 (rank : 46) | θ value | 2.52405e-10 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.466159 (rank : 47) | θ value | 4.44505e-15 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.465971 (rank : 48) | θ value | 7.34386e-10 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT6_HUMAN
|
||||||
| NC score | 0.464695 (rank : 49) | θ value | 5.62301e-10 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT4_HUMAN
|
||||||
| NC score | 0.463927 (rank : 50) | θ value | 7.34386e-10 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT5_MOUSE
|
||||||
| NC score | 0.463769 (rank : 51) | θ value | 3.41135e-07 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
SYT11_MOUSE
|
||||||
| NC score | 0.463335 (rank : 52) | θ value | 9.59137e-10 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_HUMAN
|
||||||
| NC score | 0.462802 (rank : 53) | θ value | 9.59137e-10 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT15_HUMAN
|
||||||
| NC score | 0.462011 (rank : 54) | θ value | 8.40245e-06 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
SYT4_MOUSE
|
||||||
| NC score | 0.461500 (rank : 55) | θ value | 3.64472e-09 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
SYT10_MOUSE
|
||||||
| NC score | 0.458046 (rank : 56) | θ value | 3.64472e-09 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT10_HUMAN
|
||||||
| NC score | 0.457612 (rank : 57) | θ value | 1.06045e-08 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT9_HUMAN
|
||||||
| NC score | 0.457142 (rank : 58) | θ value | 9.59137e-10 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYT9_MOUSE
|
||||||
| NC score | 0.455174 (rank : 59) | θ value | 1.38499e-08 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.449884 (rank : 60) | θ value | 5.43371e-13 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RIMS4_MOUSE
|
||||||
| NC score | 0.449171 (rank : 61) | θ value | 2.98157e-11 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| NC score | 0.447776 (rank : 62) | θ value | 3.89403e-11 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.440344 (rank : 63) | θ value | 2.69671e-12 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.436756 (rank : 64) | θ value | 1.2105e-12 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
SYT15_MOUSE
|
||||||
| NC score | 0.433553 (rank : 65) | θ value | 3.19293e-05 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
SYT8_MOUSE
|
||||||
| NC score | 0.427121 (rank : 66) | θ value | 0.00175202 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
RIMS3_HUMAN
|
||||||
| NC score | 0.380782 (rank : 67) | θ value | 3.41135e-07 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RIMS3_MOUSE
|
||||||
| NC score | 0.374874 (rank : 68) | θ value | 9.92553e-07 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80U57 | Gene names | Rims3, Kiaa0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.343959 (rank : 69) | θ value | 1.2105e-12 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SYT13_MOUSE
|
||||||
| NC score | 0.341954 (rank : 70) | θ value | 0.125558 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT13_HUMAN
|
||||||
| NC score | 0.335361 (rank : 71) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT14_HUMAN
|
||||||
| NC score | 0.327107 (rank : 72) | θ value | 0.62314 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.327066 (rank : 73) | θ value | 3.77169e-06 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
SYT14_MOUSE
|
||||||
| NC score | 0.326825 (rank : 74) | θ value | 1.38821 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.326164 (rank : 75) | θ value | 3.77169e-06 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.323558 (rank : 76) | θ value | 1.33837e-11 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.314357 (rank : 77) | θ value | 0.000461057 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.294297 (rank : 78) | θ value | 0.000121331 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.285459 (rank : 79) | θ value | 0.00134147 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
K0528_HUMAN
|
||||||
| NC score | 0.258516 (rank : 80) | θ value | 0.00509761 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_MOUSE
|
||||||
| NC score | 0.258404 (rank : 81) | θ value | 0.00509761 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.255712 (rank : 82) | θ value | 0.0252991 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.255631 (rank : 83) | θ value | 0.0193708 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.245182 (rank : 84) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
UN13D_HUMAN
|
||||||
| NC score | 0.239687 (rank : 85) | θ value | θ > 10 (rank : 198) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.237910 (rank : 86) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.232768 (rank : 87) | θ value | 1.06291 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.232470 (rank : 88) | θ value | 0.000461057 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
BAIP3_HUMAN
|
||||||
| NC score | 0.229865 (rank : 89) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
BAIP3_MOUSE
|
||||||
| NC score | 0.227986 (rank : 90) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.227566 (rank : 91) | θ value | 0.00228821 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.223684 (rank : 92) | θ value | 0.00102713 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.223266 (rank : 93) | θ value | 5.27518 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.211755 (rank : 94) | θ value | 0.00665767 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.209984 (rank : 95) | θ value | 0.00665767 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
DYSF_HUMAN
|
||||||
| NC score | 0.197320 (rank : 96) | θ value | 0.0431538 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.191720 (rank : 97) | θ value | 0.0961366 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
MYOF_HUMAN
|
||||||
| NC score | 0.179029 (rank : 98) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.176693 (rank : 99) | θ value | 0.163984 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.173539 (rank : 100) | θ value | 0.163984 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
TAC2N_HUMAN
|
||||||
| NC score | 0.171345 (rank : 101) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N9U0 | Gene names | MTAC2D1, C14orf47 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
TAC2N_MOUSE
|
||||||
| NC score | 0.170709 (rank : 102) | θ value | θ > 10 (rank : 196) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.168270 (rank : 103) | θ value | θ > 10 (rank : 197) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.166787 (rank : 104) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.166558 (rank : 105) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.164436 (rank : 106) | θ value | 3.19293e-05 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.163516 (rank : 107) | θ value | 0.0148317 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.161566 (rank : 108) | θ value | 1.06291 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
PA24D_MOUSE
|
||||||
| NC score | 0.160090 (rank : 109) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q50L43, Q3TJC5 | Gene names | Pla2g4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.154254 (rank : 110) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.153763 (rank : 111) | θ value | 0.0330416 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
OTOF_HUMAN
|
||||||
| NC score | 0.153717 (rank : 112) | θ value | 5.27518 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.151831 (rank : 113) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
PA24D_HUMAN
|
||||||
| NC score | 0.135471 (rank : 114) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
KPCG_MOUSE
|
||||||
| NC score | 0.135202 (rank : 115) | θ value | 7.34386e-10 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
KPCG_HUMAN
|
||||||
| NC score | 0.134052 (rank : 116) | θ value | 3.64472e-09 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
KPCA_HUMAN
|
||||||
| NC score | 0.128977 (rank : 117) | θ value | 9.59137e-10 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCA_MOUSE
|
||||||
| NC score | 0.125486 (rank : 118) | θ value | 8.11959e-09 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCB_HUMAN
|
||||||
| NC score | 0.124061 (rank : 119) | θ value | 6.87365e-08 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCB_MOUSE
|
||||||
| NC score | 0.124037 (rank : 120) | θ value | 1.99992e-07 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
PA24B_MOUSE
|
||||||
| NC score | 0.117004 (rank : 121) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
NCF1_HUMAN
|
||||||
| NC score | 0.109526 (rank : 122) | θ value | 9.29e-05 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P14598, O43842, Q2PP07, Q9BU90, Q9BXI7, Q9BXI8, Q9UDV9, Q9UMU2 | Gene names | NCF1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 1 (NCF-1) (Neutrophil NADPH oxidase factor 1) (47 kDa neutrophil oxidase factor) (p47-phox) (NCF-47K) (47 kDa autosomal chronic granulomatous disease protein) (NOXO2). | |||||
|
PA24E_MOUSE
|
||||||
| NC score | 0.107512 (rank : 123) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q50L42, Q8BX44 | Gene names | Pla2g4e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
NCF1_MOUSE
|
||||||
| NC score | 0.101927 (rank : 124) | θ value | 7.1131e-05 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q09014, O70144, Q9JI34 | Gene names | Ncf1 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 1 (NCF-1) (Neutrophil NADPH oxidase factor 1) (47 kDa neutrophil oxidase factor) (p47-phox) (NCF-47K). | |||||
|
PA24A_MOUSE
|
||||||
| NC score | 0.099865 (rank : 125) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
CPNE3_MOUSE
|
||||||
| NC score | 0.097265 (rank : 126) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BT60, Q5CZX9, Q5DU22 | Gene names | Cpne3, Kiaa0636 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE3_HUMAN
|
||||||
| NC score | 0.097106 (rank : 127) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75131, Q8IYA1 | Gene names | CPNE3, CPN3, KIAA0636 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-3 (Copine III). | |||||
|
PA24E_HUMAN
|
||||||
| NC score | 0.094751 (rank : 128) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q3MJ16, Q6ZSC0 | Gene names | PLA2G4E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
PA24A_HUMAN
|
||||||
| NC score | 0.094229 (rank : 129) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P47712 | Gene names | PLA2G4A, CPLA2, PLA2G4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
PA24B_HUMAN
|
||||||
| NC score | 0.091774 (rank : 130) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
CPNE5_MOUSE
|
||||||
| NC score | 0.088752 (rank : 131) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_HUMAN
|
||||||
| NC score | 0.088477 (rank : 132) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
SNX16_MOUSE
|
||||||
| NC score | 0.088090 (rank : 133) | θ value | 0.0193708 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C080 | Gene names | Snx16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-16. | |||||
|
SNX16_HUMAN
|
||||||
| NC score | 0.085121 (rank : 134) | θ value | 0.279714 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P57768, Q8N4U3 | Gene names | SNX16 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-16. | |||||
|
CPNE1_MOUSE
|
||||||
| NC score | 0.084898 (rank : 135) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE2_MOUSE
|
||||||
| NC score | 0.084495 (rank : 136) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P59108, Q8K1D7 | Gene names | Cpne2 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE2_HUMAN
|
||||||
| NC score | 0.084474 (rank : 137) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q96FN4, Q68D19, Q86XP9 | Gene names | CPNE2 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-2 (Copine II). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.083119 (rank : 138) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
CPNE8_HUMAN
|
||||||
| NC score | 0.082802 (rank : 139) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.080903 (rank : 140) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
CPNE8_MOUSE
|
||||||
| NC score | 0.080832 (rank : 141) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.076976 (rank : 142) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
CPNE7_HUMAN
|
||||||
| NC score | 0.075413 (rank : 143) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.074939 (rank : 144) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
PLCD1_HUMAN
|
||||||
| NC score | 0.071500 (rank : 145) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PA24F_MOUSE
|
||||||
| NC score | 0.070860 (rank : 146) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q50L41, Q80VQ8, Q80VV9 | Gene names | Pla2g4f | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.070699 (rank : 147) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
SNX18_HUMAN
|
||||||
| NC score | 0.070351 (rank : 148) | θ value | 0.00020696 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96RF0 | Gene names | SNAG1, SNX18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
CPNE1_HUMAN
|
||||||
| NC score | 0.070196 (rank : 149) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99829, Q6IBL3, Q9H243, Q9NTZ7 | Gene names | CPNE1, CPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
MELPH_HUMAN
|
||||||
| NC score | 0.069092 (rank : 150) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BV36, Q9HA71 | Gene names | MLPH, SLAC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Synaptotagmin-like protein 2a) (Slp homolog lacking C2 domains a). | |||||
|
SNX18_MOUSE
|
||||||
| NC score | 0.066900 (rank : 151) | θ value | 0.00020696 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91ZR2 | Gene names | Snag1, Snx18 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-18 (Sorting nexin-associated Golgi protein 1). | |||||
|
WWC1_MOUSE
|
||||||
| NC score | 0.066312 (rank : 152) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5SXA9, Q571D0, Q8K1Y3, Q8VD17, Q922W3 | Gene names | Wwc1, Kiaa0869 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-containing protein 1 (Kidney and brain protein) (KIBRA). | |||||
|
NCF4_HUMAN
|
||||||
| NC score | 0.063844 (rank : 153) | θ value | 0.0563607 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15080, O60808, Q86U56, Q9BU98, Q9NP45 | Gene names | NCF4 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
SNX22_HUMAN
|
||||||
| NC score | 0.063674 (rank : 154) | θ value | 0.0148317 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96L94, Q9H844 | Gene names | SNX22 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-22. | |||||
|
RFIP1_MOUSE
|
||||||
| NC score | 0.062919 (rank : 155) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9D620, Q3UBC2, Q8BN24 | Gene names | Rab11fip1, Rcp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
CPNE6_HUMAN
|
||||||
| NC score | 0.062733 (rank : 156) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O95741, Q53HA6, Q8WVG1 | Gene names | CPNE6 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.059421 (rank : 157) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
CPNE6_MOUSE
|
||||||
| NC score | 0.059058 (rank : 158) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Z140 | Gene names | Cpne6 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.058453 (rank : 159) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.058413 (rank : 160) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.058354 (rank : 161) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.057959 (rank : 162) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
RFIP1_HUMAN
|
||||||
| NC score | 0.057415 (rank : 163) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6WKZ4, Q6AZK4, Q6WKZ2, Q6WKZ6, Q86YV4, Q8TDL1, Q9H642 | Gene names | RAB11FIP1, RCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 1 (Rab11-FIP1) (Rab-coupling protein). | |||||
|
PA24F_HUMAN
|
||||||
| NC score | 0.056152 (rank : 164) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q68DD2, Q6ZMC8 | Gene names | PLA2G4F | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 zeta (EC 3.1.1.4) (cPLA2-zeta) (Phospholipase A2 group IVF). | |||||
|
SNX24_HUMAN
|
||||||
| NC score | 0.054762 (rank : 165) | θ value | 0.813845 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y343, Q6UY33 | Gene names | SNX24 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-24. | |||||
|
NOXO1_MOUSE
|
||||||
| NC score | 0.054649 (rank : 166) | θ value | 0.813845 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8VCM2, Q3TZA4, Q8BH41, Q9D747 | Gene names | Noxo1, Snx28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase organizer 1 (Nox organizer 1) (Sorting nexin-28). | |||||
|
SNX24_MOUSE
|
||||||
| NC score | 0.054170 (rank : 167) | θ value | 0.813845 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CRB0 | Gene names | Snx24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-24. | |||||
|
CPNE4_HUMAN
|
||||||
| NC score | 0.052457 (rank : 168) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
NCF4_MOUSE
|
||||||
| NC score | 0.052410 (rank : 169) | θ value | 0.0148317 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97369 | Gene names | Ncf4 | |||
|
Domain Architecture |

|
|||||
| Description | Neutrophil cytosol factor 4 (NCF-4) (Neutrophil NADPH oxidase factor 4) (p40-phox) (p40phox). | |||||
|
CPNE4_MOUSE
|
||||||
| NC score | 0.051008 (rank : 170) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
RASA1_HUMAN
|
||||||
| NC score | 0.043704 (rank : 171) | θ value | 0.21417 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
SGK3_MOUSE
|
||||||
| NC score | 0.027701 (rank : 172) | θ value | 0.0961366 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ERE3, Q3UAY2 | Gene names | Sgk3, Cisk, Sgkl | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Sgk3 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 3) (Serum/glucocorticoid- regulated kinase-like) (Cytokine-independent survival kinase). | |||||
|
SNX14_HUMAN
|
||||||
| NC score | 0.027665 (rank : 173) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y5W7, Q9BSD1 | Gene names | SNX14 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-14. | |||||
|
SGK3_HUMAN
|
||||||
| NC score | 0.027630 (rank : 174) | θ value | 0.125558 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 877 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96BR1, Q9P1Q7, Q9UKG5 | Gene names | SGK3, SGKL | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Sgk3 (EC 2.7.11.1) (Serum/glucocorticoid-regulated kinase 3) (Serum/glucocorticoid- regulated kinase-like). | |||||
|
SNX14_MOUSE
|
||||||
| NC score | 0.026517 (rank : 175) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BHY8 | Gene names | Snx14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-14. | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.019297 (rank : 176) | θ value | 0.813845 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.014690 (rank : 177) | θ value | 1.06291 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.013569 (rank : 178) | θ value | 0.0961366 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.013230 (rank : 179) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.013031 (rank : 180) | θ value | 1.81305 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
IQEC2_MOUSE
|
||||||
| NC score | 0.012171 (rank : 181) | θ value | 2.36792 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5DU25 | Gene names | Iqsec2, Kiaa0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
HHIP_MOUSE
|
||||||
| NC score | 0.011834 (rank : 182) | θ value | 1.81305 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TN16, Q8C0B0, Q9WU59 | Gene names | Hhip, Hip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
IQEC2_HUMAN
|
||||||
| NC score | 0.011821 (rank : 183) | θ value | 2.36792 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5JU85, O60275 | Gene names | IQSEC2, KIAA0522 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IQ motif and Sec7 domain-containing protein 2. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.011424 (rank : 184) | θ value | 2.36792 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
HHIP_HUMAN
|
||||||
| NC score | 0.010481 (rank : 185) | θ value | 2.36792 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QV1, Q6PK09, Q8NCI7, Q9BXK3, Q9H1J4, Q9H7E7 | Gene names | HHIP, HIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hedgehog-interacting protein precursor (HHIP) (HIP). | |||||
|
KKCC2_MOUSE
|
||||||
| NC score | 0.010120 (rank : 186) | θ value | 0.813845 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C078, Q80TS0, Q8BXM8, Q8C0G3, Q8CH42, Q8QZT7 | Gene names | Camkk2, Kiaa0787 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase kinase 2 (EC 2.7.11.17) (Calcium/calmodulin-dependent protein kinase kinase beta) (CaM-kinase kinase beta) (CaM-KK beta) (CaMKK beta). | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.010008 (rank : 187) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.008967 (rank : 188) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
LPIN1_HUMAN
|
||||||
| NC score | 0.004210 (rank : 189) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14693 | Gene names | LPIN1, KIAA0188 | |||
|
Domain Architecture |

|
|||||
| Description | Lipin-1. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.002866 (rank : 190) | θ value | 1.81305 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
ABCG4_HUMAN
|
||||||
| NC score | 0.002345 (rank : 191) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H172 | Gene names | ABCG4, WHITE2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-binding cassette sub-family G member 4. | |||||
|
ABCAD_MOUSE
|
||||||
| NC score | 0.001816 (rank : 192) | θ value | 5.27518 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.000739 (rank : 193) | θ value | 5.27518 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.000273 (rank : 194) | θ value | 6.88961 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CAC1B_HUMAN
|
||||||
| NC score | 0.000217 (rank : 195) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00975 | Gene names | CACNA1B, CACH5, CACNL1A5 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | -0.003384 (rank : 196) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
OPSG_MOUSE
|
||||||
| NC score | -0.004917 (rank : 197) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35599, Q548Z4 | Gene names | Opn1mw, Gcp | |||
|
Domain Architecture |

|
|||||
| Description | Green-sensitive opsin (Green cone photoreceptor pigment) (Medium wavelength-sensitive cone opsin) (M opsin). | |||||
|
AIOL_HUMAN
|
||||||
| NC score | -0.006912 (rank : 198) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKT9, Q8WWQ9, Q8WWR0, Q8WWR1, Q8WWR2, Q8WWR3 | Gene names | IKZF3, ZNFN1A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||