Please be patient as the page loads
|
PLCD1_HUMAN
|
||||||
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLCD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PLCD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989088 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PLCB2_HUMAN
|
||||||
| θ value | 2.91998e-83 (rank : 3) | NC score | 0.756920 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 4.21644e-82 (rank : 4) | NC score | 0.743039 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 3.56943e-81 (rank : 5) | NC score | 0.753636 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PLCG2_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 6) | NC score | 0.625746 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
PLCG1_MOUSE
|
||||||
| θ value | 2.18062e-54 (rank : 7) | NC score | 0.611318 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
PLCG1_HUMAN
|
||||||
| θ value | 1.08223e-53 (rank : 8) | NC score | 0.617079 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
PLCB4_HUMAN
|
||||||
| θ value | 1.788e-48 (rank : 9) | NC score | 0.655236 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
PLCB3_HUMAN
|
||||||
| θ value | 3.98324e-48 (rank : 10) | NC score | 0.748116 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 8.87376e-48 (rank : 11) | NC score | 0.778129 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
SYT1_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 12) | NC score | 0.195491 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |
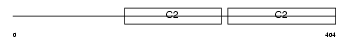
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 13) | NC score | 0.195486 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |
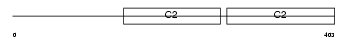
|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 0.000461057 (rank : 14) | NC score | 0.131749 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 15) | NC score | 0.130623 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
SYT7_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 16) | NC score | 0.193260 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |
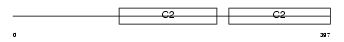
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT7_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 17) | NC score | 0.193353 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |
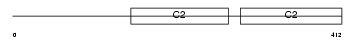
|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
CPNE6_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.112916 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Z140 | Gene names | Cpne6 | |||
|
Domain Architecture |
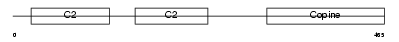
|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
CPNE6_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 19) | NC score | 0.110007 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95741, Q53HA6, Q8WVG1 | Gene names | CPNE6 | |||
|
Domain Architecture |
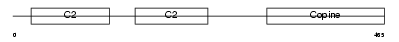
|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
SEGN_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 20) | NC score | 0.094152 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
SYT5_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 21) | NC score | 0.188725 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |
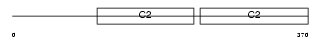
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT11_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 22) | NC score | 0.181801 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |
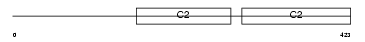
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 23) | NC score | 0.181803 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |
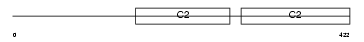
|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
NEDD4_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.116997 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
SYT5_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 25) | NC score | 0.188036 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |
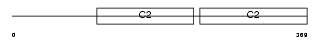
|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
RASL2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 26) | NC score | 0.164242 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
AB2BP_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 27) | NC score | 0.112551 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D6J4, Q3TYZ9, Q9ESR0 | Gene names | Apba2bp, Xb51 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 protein-binding family A member 2-binding protein (X11L-binding protein 51) (mXB51). | |||||
|
DOC2B_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 28) | NC score | 0.200412 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 29) | NC score | 0.114560 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
DOC2B_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 30) | NC score | 0.199983 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
SEGN_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 31) | NC score | 0.081930 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |
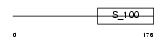
|
|||||
| Description | Secretagogin. | |||||
|
SYT2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 32) | NC score | 0.184312 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |
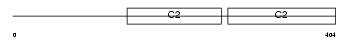
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
AB2BP_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 33) | NC score | 0.105803 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P71, Q5JWF6, Q5JWF7, Q86VV1, Q9H433, Q9H8G8, Q9HBW7, Q9HCQ9 | Gene names | APBA2BP, NECAB3, NIP1, XB51 | |||
|
Domain Architecture |
|
|||||
| Description | Amyloid beta A4 protein-binding family A member 2-binding protein (Neuronal calcium-binding protein 3) (NECAB 3) (X11L-binding protein 51) (Nek2-interacting protein 1). | |||||
|
CALU_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 34) | NC score | 0.072908 (rank : 111) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 35) | NC score | 0.130611 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
KPCG_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 36) | NC score | 0.057352 (rank : 134) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
SYT2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 37) | NC score | 0.183638 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
KPCA_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 38) | NC score | 0.054210 (rank : 142) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCG_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 39) | NC score | 0.057690 (rank : 132) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
DOC2A_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 40) | NC score | 0.210204 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
MYOF_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 41) | NC score | 0.141288 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
CALU_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 42) | NC score | 0.068993 (rank : 118) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
DOC2A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 43) | NC score | 0.206067 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
KPCA_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 44) | NC score | 0.052609 (rank : 150) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
SYT3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 45) | NC score | 0.173379 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.173200 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
K0528_HUMAN
|
||||||
| θ value | 0.163984 (rank : 47) | NC score | 0.157497 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_MOUSE
|
||||||
| θ value | 0.163984 (rank : 48) | NC score | 0.157216 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
RASL1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.135709 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
KPCB_HUMAN
|
||||||
| θ value | 0.21417 (rank : 50) | NC score | 0.052485 (rank : 151) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
KPCB_MOUSE
|
||||||
| θ value | 0.21417 (rank : 51) | NC score | 0.053154 (rank : 148) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 52) | NC score | 0.172219 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | 0.21417 (rank : 53) | NC score | 0.171811 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
CAB45_MOUSE
|
||||||
| θ value | 0.279714 (rank : 54) | NC score | 0.049706 (rank : 164) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61112, Q61113 | Gene names | Sdf4, Cab45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
CPNE7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 55) | NC score | 0.089920 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
MCM8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 56) | NC score | 0.031181 (rank : 170) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
PA24D_MOUSE
|
||||||
| θ value | 0.365318 (rank : 57) | NC score | 0.115717 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q50L43, Q3TJC5 | Gene names | Pla2g4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
BAIP3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 58) | NC score | 0.146076 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
CU013_HUMAN
|
||||||
| θ value | 0.47712 (rank : 59) | NC score | 0.045270 (rank : 165) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 60) | NC score | 0.025827 (rank : 173) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
SYT4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 61) | NC score | 0.170822 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
CPNE2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 62) | NC score | 0.086164 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59108, Q8K1D7 | Gene names | Cpne2 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-2 (Copine II). | |||||
|
SYT4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 63) | NC score | 0.168679 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |
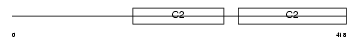
|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
BAIP3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 64) | NC score | 0.141602 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
CPNE2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 65) | NC score | 0.087010 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96FN4, Q68D19, Q86XP9 | Gene names | CPNE2 | |||
|
Domain Architecture |
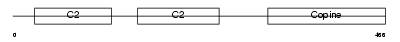
|
|||||
| Description | Copine-2 (Copine II). | |||||
|
PLSI_HUMAN
|
||||||
| θ value | 1.38821 (rank : 66) | NC score | 0.020308 (rank : 174) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.018718 (rank : 176) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
SYT9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.161433 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.094518 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SYT9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.161038 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |
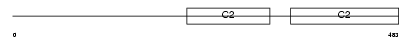
|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
UN13D_HUMAN
|
||||||
| θ value | 2.36792 (rank : 71) | NC score | 0.135838 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
CYLD_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.028344 (rank : 171) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
DYSF_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.110793 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
MCM8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.019431 (rank : 175) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.097721 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TEX15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.037782 (rank : 167) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
CAB45_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.040561 (rank : 166) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |
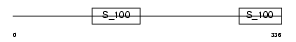
|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
CALB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.034771 (rank : 168) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CYLD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.027177 (rank : 172) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQC7, O94934, Q7L3N6, Q96EH0, Q9NZX9 | Gene names | CYLD, CYLD1, KIAA0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
TEC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.016874 (rank : 177) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42680 | Gene names | TEC, PSCTK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
DOC2G_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.170290 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.013567 (rank : 181) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
LX12E_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.005980 (rank : 184) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
NSF1C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.012543 (rank : 182) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZ44, Q80Y15, Q8BYL3 | Gene names | Nsfl1c | |||
|
Domain Architecture |
|
|||||
| Description | NSFL1 cofactor p47 (p97 cofactor p47). | |||||
|
SYT10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.157324 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.137192 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
TEC_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.016382 (rank : 179) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24604, Q9R1M9, Q9WVN0, Q9WVN1, Q9WVN2, Q9WVN3 | Gene names | Tec | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
TF2LY_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.016639 (rank : 178) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUE0 | Gene names | TGIF2LY, TGIFLY | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein TGIF2LY (TGFB-induced factor 2-like protein, Y- linked) (TGF(beta)induced transcription factor 2-like protein) (TGIF- like on the Y). | |||||
|
CALB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.031741 (rank : 169) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
CPNE8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.081956 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
EFHC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.012279 (rank : 183) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JVL4, Q5XKM4, Q6E1U7, Q6E1U8, Q8WUL2, Q9NVW6 | Gene names | EFHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.016089 (rank : 180) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
TGFB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.003354 (rank : 185) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27090 | Gene names | Tgfb2 | |||
|
Domain Architecture |
|
|||||
| Description | Transforming growth factor beta-2 precursor (TGF-beta-2). | |||||
|
APS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.053981 (rank : 144) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14492 | Gene names | APS | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
CPNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.071450 (rank : 114) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99829, Q6IBL3, Q9H243, Q9NTZ7 | Gene names | CPNE1, CPN1 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.079170 (rank : 99) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.085067 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75131, Q8IYA1 | Gene names | CPNE3, CPN3, KIAA0636 | |||
|
Domain Architecture |
|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.086135 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BT60, Q5CZX9, Q5DU22 | Gene names | Cpne3, Kiaa0636 | |||
|
Domain Architecture |
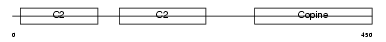
|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.067210 (rank : 121) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |
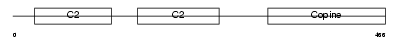
|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.066980 (rank : 122) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
CPNE5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.081611 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.081900 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |
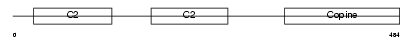
|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.078753 (rank : 100) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |
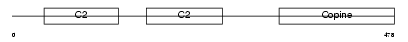
|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
CRK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053408 (rank : 147) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46108 | Gene names | CRK | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
CRK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.052240 (rank : 153) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64010 | Gene names | Crk, Crko | |||
|
Domain Architecture |
|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
DAPP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.060246 (rank : 129) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
DAPP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.065341 (rank : 124) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |
|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.118789 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.120326 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
GRAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.052060 (rank : 154) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
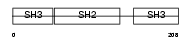
Domain Architecture |
|
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRAP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.053646 (rank : 145) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.052410 (rank : 152) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62993, P29354, Q14450, Q63057, Q63059 | Gene names | GRB2, ASH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash). | |||||
|
GRB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053473 (rank : 146) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60631, Q61240 | Gene names | Grb2 | |||
|
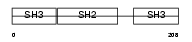
Domain Architecture |
|
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2). | |||||
|
HSH2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.053083 (rank : 149) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.073864 (rank : 108) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.070450 (rank : 115) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
ITSN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.073850 (rank : 109) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.077426 (rank : 101) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.057469 (rank : 133) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
LCP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.074448 (rank : 107) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
OTOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.092895 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
OTOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.091664 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.054675 (rank : 140) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.054199 (rank : 143) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.071500 (rank : 113) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
P55G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.054395 (rank : 141) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P55G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050361 (rank : 162) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
P85B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.051372 (rank : 157) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
PA24A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.066175 (rank : 123) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
PA24B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.056855 (rank : 135) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PA24B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.068568 (rank : 119) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PA24D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.069723 (rank : 117) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
PA24E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051877 (rank : 156) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3MJ16, Q6ZSC0 | Gene names | PLA2G4E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
PA24E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.059861 (rank : 130) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q50L42, Q8BX44 | Gene names | Pla2g4e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
RASA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.069800 (rank : 116) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.074749 (rank : 106) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.076952 (rank : 103) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.076654 (rank : 104) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.065231 (rank : 125) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RASL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.126670 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.077283 (rank : 102) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
RIMS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.081382 (rank : 96) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
RIMS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.083801 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.082578 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
RIMS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051345 (rank : 158) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
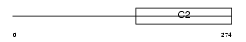
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.062150 (rank : 128) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
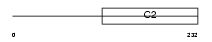
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.062704 (rank : 127) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |
|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
SH22A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.072762 (rank : 112) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
SH22A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.064187 (rank : 126) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
SH24A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.067787 (rank : 120) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
SHE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050540 (rank : 161) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VZ18, Q8TEQ5 | Gene names | SHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
SHE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051034 (rank : 159) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BSD5, Q3TZT0 | Gene names | She | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
SMUF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.055956 (rank : 138) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.055749 (rank : 139) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.056742 (rank : 136) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
SYT10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.157410 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.134306 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.126837 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.124639 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYT14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.118049 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYT14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.120809 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
SYT15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.136992 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
SYT15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.132827 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
SYT6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.153657 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.154139 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |
|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.138968 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.118221 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.109827 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.114400 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.112644 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.104250 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
SYTL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.123029 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.122023 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.118068 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYTL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.115448 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
TAC2N_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.074911 (rank : 105) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N9U0 | Gene names | MTAC2D1, C14orf47 | |||
|
Domain Architecture |
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
TAC2N_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.073375 (rank : 110) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.081004 (rank : 98) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.081156 (rank : 97) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.087116 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
VAV2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.052022 (rank : 155) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
VAV3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.058519 (rank : 131) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.055972 (rank : 137) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
VAV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.050242 (rank : 163) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
VAV_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.050764 (rank : 160) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
PLCD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | P51178 | Gene names | PLCD1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PLCD1_MOUSE
|
||||||
| NC score | 0.989088 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8R3B1 | Gene names | Plcd1, Plcd | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase delta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-delta-1) (Phospholipase C-delta-1) (PLC-III). | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.778129 (rank : 3) | θ value | 8.87376e-48 (rank : 11) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PLCB2_HUMAN
|
||||||
| NC score | 0.756920 (rank : 4) | θ value | 2.91998e-83 (rank : 3) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q00722 | Gene names | PLCB2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-2) (PLC-beta-2). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.753636 (rank : 5) | θ value | 3.56943e-81 (rank : 5) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PLCB3_HUMAN
|
||||||
| NC score | 0.748116 (rank : 6) | θ value | 3.98324e-48 (rank : 10) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.743039 (rank : 7) | θ value | 4.21644e-82 (rank : 4) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PLCB4_HUMAN
|
||||||
| NC score | 0.655236 (rank : 8) | θ value | 1.788e-48 (rank : 9) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
PLCG2_HUMAN
|
||||||
| NC score | 0.625746 (rank : 9) | θ value | 1.96772e-63 (rank : 6) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P16885, Q969T5 | Gene names | PLCG2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 2 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-2) (Phospholipase C-gamma-2) (PLC-IV). | |||||
|
PLCG1_HUMAN
|
||||||
| NC score | 0.617079 (rank : 10) | θ value | 1.08223e-53 (rank : 8) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19174, Q2V575 | Gene names | PLCG1, PLC1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
PLCG1_MOUSE
|
||||||
| NC score | 0.611318 (rank : 11) | θ value | 2.18062e-54 (rank : 7) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q62077, Q6P1G1 | Gene names | Plcg1, Plcg-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (PLC-gamma-1) (Phospholipase C-gamma-1) (PLC-II) (PLC-148). | |||||
|
DOC2A_MOUSE
|
||||||
| NC score | 0.210204 (rank : 12) | θ value | 0.0330416 (rank : 40) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.206067 (rank : 13) | θ value | 0.0563607 (rank : 43) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
DOC2B_MOUSE
|
||||||
| NC score | 0.200412 (rank : 14) | θ value | 0.00869519 (rank : 28) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2B_HUMAN
|
||||||
| NC score | 0.199983 (rank : 15) | θ value | 0.0113563 (rank : 30) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
SYT1_HUMAN
|
||||||
| NC score | 0.195491 (rank : 16) | θ value | 0.000270298 (rank : 12) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P21579 | Gene names | SYT1, SYT | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT1_MOUSE
|
||||||
| NC score | 0.195486 (rank : 17) | θ value | 0.000270298 (rank : 13) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P46096 | Gene names | Syt1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-1 (Synaptotagmin I) (SytI) (p65). | |||||
|
SYT7_HUMAN
|
||||||
| NC score | 0.193353 (rank : 18) | θ value | 0.000602161 (rank : 17) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43581 | Gene names | SYT7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT7_MOUSE
|
||||||
| NC score | 0.193260 (rank : 19) | θ value | 0.000461057 (rank : 16) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R0N7 | Gene names | Syt7 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-7 (Synaptotagmin VII) (SytVII). | |||||
|
SYT5_HUMAN
|
||||||
| NC score | 0.188725 (rank : 20) | θ value | 0.00228821 (rank : 21) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O00445, Q86X72 | Gene names | SYT5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV). | |||||
|
SYT5_MOUSE
|
||||||
| NC score | 0.188036 (rank : 21) | θ value | 0.00509761 (rank : 25) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9R0N5 | Gene names | Syt5, Syt9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-5 (Synaptotagmin V) (SytV) (Synaptotagmin IX). | |||||
|
SYT2_MOUSE
|
||||||
| NC score | 0.184312 (rank : 22) | θ value | 0.0113563 (rank : 32) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P46097, Q8R0E1 | Gene names | Syt2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT2_HUMAN
|
||||||
| NC score | 0.183638 (rank : 23) | θ value | 0.0193708 (rank : 37) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8N9I0, Q496K5, Q8NBE5 | Gene names | SYT2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-2 (Synaptotagmin II) (SytII). | |||||
|
SYT11_MOUSE
|
||||||
| NC score | 0.181803 (rank : 24) | θ value | 0.00298849 (rank : 23) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9R0N3 | Gene names | Syt11 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT11_HUMAN
|
||||||
| NC score | 0.181801 (rank : 25) | θ value | 0.00298849 (rank : 22) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BT88, Q14998, Q68CT5, Q8IXU3, Q96SU2 | Gene names | SYT11, KIAA0080 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-11 (Synaptotagmin XI) (SytXI). | |||||
|
SYT3_HUMAN
|
||||||
| NC score | 0.173379 (rank : 26) | θ value | 0.0961366 (rank : 45) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQG1, Q8N5Z1, Q8N640 | Gene names | SYT3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
SYT3_MOUSE
|
||||||
| NC score | 0.173200 (rank : 27) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35681, P97791, Q80WV1 | Gene names | Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-3 (Synaptotagmin III) (SytIII). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.172219 (rank : 28) | θ value | 0.21417 (rank : 52) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.171811 (rank : 29) | θ value | 0.21417 (rank : 53) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
SYT4_HUMAN
|
||||||
| NC score | 0.170822 (rank : 30) | θ value | 0.62314 (rank : 61) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2B2, Q9P2K4 | Gene names | SYT4, KIAA1342 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
DOC2G_MOUSE
|
||||||
| NC score | 0.170290 (rank : 31) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9ESN1, Q6P8R8 | Gene names | Doc2g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein gamma (Doc2-gamma). | |||||
|
SYT4_MOUSE
|
||||||
| NC score | 0.168679 (rank : 32) | θ value | 1.06291 (rank : 63) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P40749, Q3UFC1, Q8BGH3, Q8BRL6 | Gene names | Syt4, Syt3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-4 (Synaptotagmin IV) (SytIV). | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.164242 (rank : 33) | θ value | 0.00665767 (rank : 26) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
SYT9_HUMAN
|
||||||
| NC score | 0.161433 (rank : 34) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q86SS6 | Gene names | SYT9 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX). | |||||
|
SYT9_MOUSE
|
||||||
| NC score | 0.161038 (rank : 35) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
K0528_HUMAN
|
||||||
| NC score | 0.157497 (rank : 36) | θ value | 0.163984 (rank : 47) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
SYT10_HUMAN
|
||||||
| NC score | 0.157410 (rank : 37) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6XYQ8 | Gene names | SYT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
SYT10_MOUSE
|
||||||
| NC score | 0.157324 (rank : 38) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9R0N4 | Gene names | Syt10 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-10 (Synaptotagmin X) (SytX). | |||||
|
K0528_MOUSE
|
||||||
| NC score | 0.157216 (rank : 39) | θ value | 0.163984 (rank : 48) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
SYT6_MOUSE
|
||||||
| NC score | 0.154139 (rank : 40) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9R0N8 | Gene names | Syt6 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
SYT6_HUMAN
|
||||||
| NC score | 0.153657 (rank : 41) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5T7P8 | Gene names | SYT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-6 (Synaptotagmin VI) (SytVI). | |||||
|
BAIP3_HUMAN
|
||||||
| NC score | 0.146076 (rank : 42) | θ value | 0.47712 (rank : 58) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
BAIP3_MOUSE
|
||||||
| NC score | 0.141602 (rank : 43) | θ value | 1.38821 (rank : 64) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80TT2, Q6RUT4 | Gene names | Baiap3, Gm937, Kiaa0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
MYOF_HUMAN
|
||||||
| NC score | 0.141288 (rank : 44) | θ value | 0.0330416 (rank : 41) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
SYT8_MOUSE
|
||||||
| NC score | 0.138968 (rank : 45) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9R0N6, Q64366, Q8C6E7 | Gene names | Syt8 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-8 (Synaptotagmin VIII) (SytVIII). | |||||
|
SYT12_MOUSE
|
||||||
| NC score | 0.137192 (rank : 46) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q920N7 | Gene names | Syt12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT15_HUMAN
|
||||||
| NC score | 0.136992 (rank : 47) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BQS2, Q7Z439, Q7Z440 | Gene names | SYT15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV) (Chr10Syt). | |||||
|
UN13D_HUMAN
|
||||||
| NC score | 0.135838 (rank : 48) | θ value | 2.36792 (rank : 71) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.135709 (rank : 49) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
SYT12_HUMAN
|
||||||
| NC score | 0.134306 (rank : 50) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8IV01 | Gene names | SYT12 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-12 (Synaptotagmin XII) (SytXII). | |||||
|
SYT15_MOUSE
|
||||||
| NC score | 0.132827 (rank : 51) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8C6N3, Q7TN81, Q7TN82, Q8C999 | Gene names | Syt15 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-15 (Synaptotagmin XV) (SytXV). | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.131749 (rank : 52) | θ value | 0.000461057 (rank : 14) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
NED4L_MOUSE
|
||||||
| NC score | 0.130623 (rank : 53) | θ value | 0.000461057 (rank : 15) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CFI0, Q8BRT9, Q8BS42, Q99PK2 | Gene names | Nedd4l, Kiaa0439, Nedd4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.130611 (rank : 54) | θ value | 0.0193708 (rank : 35) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
SYT13_HUMAN
|
||||||
| NC score | 0.126837 (rank : 55) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7L8C5, Q9BQS3, Q9H041, Q9P2C0 | Gene names | SYT13, KIAA1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.126670 (rank : 56) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
SYT13_MOUSE
|
||||||
| NC score | 0.124639 (rank : 57) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9EQT6, Q6ZPR2, Q8BRK6 | Gene names | Syt13, Kiaa1427 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-13 (Synaptotagmin XIII) (SytXIII). | |||||
|
SYTL4_HUMAN
|
||||||
| NC score | 0.123029 (rank : 58) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96C24, Q5H9J3, Q5JPG8, Q8N9P4, Q9H4R0, Q9H4R1 | Gene names | SYTL4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYTL4_MOUSE
|
||||||
| NC score | 0.122023 (rank : 59) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9R0Q1, Q8R321, Q9R0Q0 | Gene names | Sytl4, Slp4 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 4 (Exophilin-2) (Granuphilin). | |||||
|
SYT14_MOUSE
|
||||||
| NC score | 0.120809 (rank : 60) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q7TN84 | Gene names | Syt14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.120326 (rank : 61) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.118789 (rank : 62) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.118221 (rank : 63) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
SYTL5_HUMAN
|
||||||
| NC score | 0.118068 (rank : 64) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8TDW5 | Gene names | SYTL5, SLP5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
SYT14_HUMAN
|
||||||
| NC score | 0.118049 (rank : 65) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8NB59, Q5THX7, Q707N3, Q707N4, Q707N5, Q707N6, Q707N7 | Gene names | SYT14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptotagmin-14 (Synaptotagmin XIV) (SytXIV). | |||||
|
NEDD4_MOUSE
|
||||||
| NC score | 0.116997 (rank : 66) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46935, O08758, Q3UZI2, Q8BGB3 | Gene names | Nedd4, Kiaa0093, Nedd-4, Nedd4a | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-) (Neural precursor cell expressed developmentally down-regulated protein 4). | |||||
|
PA24D_MOUSE
|
||||||
| NC score | 0.115717 (rank : 67) | θ value | 0.365318 (rank : 57) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q50L43, Q3TJC5 | Gene names | Pla2g4d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
SYTL5_MOUSE
|
||||||
| NC score | 0.115448 (rank : 68) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q80T23, Q812E5 | Gene names | Sytl5, Slp5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 5. | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.114560 (rank : 69) | θ value | 0.00869519 (rank : 29) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.114400 (rank : 70) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
CPNE6_MOUSE
|
||||||
| NC score | 0.112916 (rank : 71) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9Z140 | Gene names | Cpne6 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
SYTL2_MOUSE
|
||||||
| NC score | 0.112644 (rank : 72) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 253 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q99N50, Q8BT37, Q99J89, Q99J90, Q99N51, Q99N52, Q99N55, Q99N56 | Gene names | Sytl2, Slp2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
AB2BP_MOUSE
|
||||||
| NC score | 0.112551 (rank : 73) | θ value | 0.00869519 (rank : 27) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D6J4, Q3TYZ9, Q9ESR0 | Gene names | Apba2bp, Xb51 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 protein-binding family A member 2-binding protein (X11L-binding protein 51) (mXB51). | |||||
|
DYSF_HUMAN
|
||||||
| NC score | 0.110793 (rank : 74) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
CPNE6_HUMAN
|
||||||
| NC score | 0.110007 (rank : 75) | θ value | 0.00228821 (rank : 19) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O95741, Q53HA6, Q8WVG1 | Gene names | CPNE6 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-6 (Copine VI) (Neuronal-copine) (N-copine). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.109827 (rank : 76) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
AB2BP_HUMAN
|
||||||
| NC score | 0.105803 (rank : 77) | θ value | 0.0148317 (rank : 33) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P71, Q5JWF6, Q5JWF7, Q86VV1, Q9H433, Q9H8G8, Q9HBW7, Q9HCQ9 | Gene names | APBA2BP, NECAB3, NIP1, XB51 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 protein-binding family A member 2-binding protein (Neuronal calcium-binding protein 3) (NECAB 3) (X11L-binding protein 51) (Nek2-interacting protein 1). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.104250 (rank : 78) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.097721 (rank : 79) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.094518 (rank : 80) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SEGN_MOUSE
|
||||||
| NC score | 0.094152 (rank : 81) | θ value | 0.00228821 (rank : 20) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91WD9 | Gene names | Scgn | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
OTOF_HUMAN
|
||||||
| NC score | 0.092895 (rank : 82) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.091664 (rank : 83) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
CPNE7_HUMAN
|
||||||
| NC score | 0.089920 (rank : 84) | θ value | 0.279714 (rank : 55) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UBL6 | Gene names | CPNE7 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-7 (Copine VII). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.087116 (rank : 85) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
CPNE2_HUMAN
|
||||||
| NC score | 0.087010 (rank : 86) | θ value | 1.38821 (rank : 65) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96FN4, Q68D19, Q86XP9 | Gene names | CPNE2 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE2_MOUSE
|
||||||
| NC score | 0.086164 (rank : 87) | θ value | 1.06291 (rank : 62) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59108, Q8K1D7 | Gene names | Cpne2 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-2 (Copine II). | |||||
|
CPNE3_MOUSE
|
||||||
| NC score | 0.086135 (rank : 88) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BT60, Q5CZX9, Q5DU22 | Gene names | Cpne3, Kiaa0636 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-3 (Copine III). | |||||
|
CPNE3_HUMAN
|
||||||
| NC score | 0.085067 (rank : 89) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O75131, Q8IYA1 | Gene names | CPNE3, CPN3, KIAA0636 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-3 (Copine III). | |||||
|
RIMS2_HUMAN
|
||||||
| NC score | 0.083801 (rank : 90) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9UQ26, O43413, Q86XL9, Q8IWV9, Q8IWW1 | Gene names | RIMS2, KIAA0751, RIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2). | |||||
|
RIMS2_MOUSE
|
||||||
| NC score | 0.082578 (rank : 91) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9EQZ7, Q8C433, Q8CCK2 | Gene names | Rims2, Rab3ip2, Rim2 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 2 (Rab3-interacting molecule 2) (RIM 2) (Rab3-interacting protein 2). | |||||
|
CPNE8_HUMAN
|
||||||
| NC score | 0.081956 (rank : 92) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q86YQ8, Q2TB41 | Gene names | CPNE8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
SEGN_HUMAN
|
||||||
| NC score | 0.081930 (rank : 93) | θ value | 0.0113563 (rank : 31) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O76038, Q5VV44, Q96QV7, Q9UJF6 | Gene names | SCGN, SECRET | |||
|
Domain Architecture |

|
|||||
| Description | Secretagogin. | |||||
|
CPNE5_MOUSE
|
||||||
| NC score | 0.081900 (rank : 94) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8JZW4 | Gene names | Cpne5 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
CPNE5_HUMAN
|
||||||
| NC score | 0.081611 (rank : 95) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9HCH3 | Gene names | CPNE5, KIAA1599 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-5 (Copine V). | |||||
|
RIMS1_HUMAN
|
||||||
| NC score | 0.081382 (rank : 96) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86UR5, O15048, Q8TDY9, Q8TDZ5, Q9HBA1, Q9HBA2, Q9HBA3, Q9HBA4, Q9HBA5, Q9HBA6 | Gene names | RIMS1, KIAA0340, RIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 1 (Rab3-interacting molecule 1) (RIM 1). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.081156 (rank : 97) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.081004 (rank : 98) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
CPNE1_MOUSE
|
||||||
| NC score | 0.079170 (rank : 99) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE8_MOUSE
|
||||||
| NC score | 0.078753 (rank : 100) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DC53, Q9CUZ8 | Gene names | Cpne8 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-8 (Copine VIII). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.077426 (rank : 101) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.077283 (rank : 102) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.076952 (rank : 103) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.076654 (rank : 104) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
TAC2N_HUMAN
|
||||||
| NC score | 0.074911 (rank : 105) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N9U0 | Gene names | MTAC2D1, C14orf47 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.074749 (rank : 106) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
LCP2_MOUSE
|
||||||
| NC score | 0.074448 (rank : 107) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60787 | Gene names | Lcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.073864 (rank : 108) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN2_HUMAN
|
||||||
| NC score | 0.073850 (rank : 109) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1173 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NZM3, O95062, Q15812, Q9HAK4, Q9NXE6, Q9NYG0, Q9NZM2, Q9ULG4 | Gene names | ITSN2, KIAA1256, SH3D1B, SWAP | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (SH3P18) (SH3P18-like WASP-associated protein). | |||||
|
TAC2N_MOUSE
|
||||||
| NC score | 0.073375 (rank : 110) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
CALU_MOUSE
|
||||||
| NC score | 0.072908 (rank : 111) | θ value | 0.0193708 (rank : 34) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35887 | Gene names | Calu | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin). | |||||
|
SH22A_HUMAN
|
||||||
| NC score | 0.072762 (rank : 112) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NP31, O43817, Q9UPA7 | Gene names | SH2D2A, SCAP, TSAD, VRAP | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (T cell-specific adapter protein) (TSAd) (VEGF receptor-associated protein) (SH2 domain-containing adapter protein). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.071500 (rank : 113) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
CPNE1_HUMAN
|
||||||
| NC score | 0.071450 (rank : 114) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q99829, Q6IBL3, Q9H243, Q9NTZ7 | Gene names | CPNE1, CPN1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.070450 (rank : 115) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
RASA1_HUMAN
|
||||||
| NC score | 0.069800 (rank : 116) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P20936 | Gene names | RASA1, RASA | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 1 (GTPase-activating protein) (GAP) (Ras p21 protein activator) (p120GAP) (RasGAP). | |||||
|
PA24D_HUMAN
|
||||||
| NC score | 0.069723 (rank : 117) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86XP0, Q8N176 | Gene names | PLA2G4D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 delta (EC 3.1.1.4) (cPLA2-delta) (Phospholipase A2 group IVD). | |||||
|
CALU_HUMAN
|
||||||
| NC score | 0.068993 (rank : 118) | θ value | 0.0431538 (rank : 42) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43852, O60456, Q6FHB9, Q96RL3, Q9NR43 | Gene names | CALU | |||
|
Domain Architecture |

|
|||||
| Description | Calumenin precursor (Crocalbin) (IEF SSP 9302). | |||||
|
PA24B_MOUSE
|
||||||
| NC score | 0.068568 (rank : 119) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.067787 (rank : 120) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
CPNE4_HUMAN
|
||||||
| NC score | 0.067210 (rank : 121) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| NC score | 0.066980 (rank : 122) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
PA24A_MOUSE
|
||||||
| NC score | 0.066175 (rank : 123) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P47713 | Gene names | Pla2g4a, Cpla2, Pla2g4 | |||
|
Domain Architecture |

|
|||||
| Description | Cytosolic phospholipase A2 (cPLA2) (Phospholipase A2 group IVA) [Includes: Phospholipase A2 (EC 3.1.1.4) (Phosphatidylcholine 2- acylhydrolase); Lysophospholipase (EC 3.1.1.5)]. | |||||
|
DAPP1_MOUSE
|
||||||
| NC score | 0.065341 (rank : 124) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXT1, Q9R178 | Gene names | Dapp1, Bam32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (mDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.065231 (rank : 125) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
SH22A_MOUSE
|
||||||
| NC score | 0.064187 (rank : 126) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXK9, Q9JHP6 | Gene names | Sh2d2a, Lad, Ribp | |||
|
Domain Architecture |

|
|||||
| Description | SH2 domain protein 2A (Lck-associated adapter protein) (Lad) (Rlk/Itk- binding protein) (Ribp). | |||||
|
RIMS4_MOUSE
|
||||||
| NC score | 0.062704 (rank : 127) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60191 | Gene names | Rims4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
RIMS4_HUMAN
|
||||||
| NC score | 0.062150 (rank : 128) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H426, Q5JWT7 | Gene names | RIMS4 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 4 (Rab3-interacting molecule 4) (RIM 4) (RIM4 gamma). | |||||
|
DAPP1_HUMAN
|
||||||
| NC score | 0.060246 (rank : 129) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UN19, Q8TCK5, Q9UHF2 | Gene names | DAPP1, BAM32 | |||
|
Domain Architecture |

|
|||||
| Description | Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3- phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (B lymphocyte adapter protein Bam32). | |||||
|
PA24E_MOUSE
|
||||||
| NC score | 0.059861 (rank : 130) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q50L42, Q8BX44 | Gene names | Pla2g4e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
VAV3_HUMAN
|
||||||
| NC score | 0.058519 (rank : 131) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKW4, O95230, Q9Y5X8 | Gene names | VAV3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
KPCG_MOUSE
|
||||||
| NC score | 0.057690 (rank : 132) | θ value | 0.0252991 (rank : 39) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P63318, P05697 | Gene names | Prkcg, Pkcc, Pkcg, Prkcc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.057469 (rank : 133) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
KPCG_HUMAN
|
||||||
| NC score | 0.057352 (rank : 134) | θ value | 0.0193708 (rank : 36) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1126 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P05129 | Gene names | PRKCG, PKCG | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C gamma type (EC 2.7.11.13) (PKC-gamma). | |||||
|
PA24B_HUMAN
|
||||||
| NC score | 0.056855 (rank : 135) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.056742 (rank : 136) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
VAV3_MOUSE
|
||||||
| NC score | 0.055972 (rank : 137) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R0C8, Q7TS85 | Gene names | Vav3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-3. | |||||
|
SMUF1_HUMAN
|
||||||
| NC score | 0.055956 (rank : 138) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9HCE7, O75853, Q9UJT8 | Gene names | SMURF1, KIAA1625 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1) (hSMURF1). | |||||
|
SMUF1_MOUSE
|
||||||
| NC score | 0.055749 (rank : 139) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9CUN6, Q3U412, Q8K300 | Gene names | Smurf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Smad ubiquitination regulatory factor 1 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF1) (Smad-specific E3 ubiquitin ligase 1). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.054675 (rank : 140) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.054395 (rank : 141) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
KPCA_MOUSE
|
||||||
| NC score | 0.054210 (rank : 142) | θ value | 0.0252991 (rank : 38) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.054199 (rank : 143) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
APS_HUMAN
|
||||||
| NC score | 0.053981 (rank : 144) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14492 | Gene names | APS | |||
|
Domain Architecture |

|
|||||
| Description | SH2 and PH domain-containing adapter protein APS (Adapter protein with pleckstrin homology and Src homology 2 domains). | |||||
|
GRAP_MOUSE
|
||||||
| NC score | 0.053646 (rank : 145) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CX99, Q3U545 | Gene names | Grap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRB2-related adapter protein. | |||||
|
GRB2_MOUSE
|
||||||
| NC score | 0.053473 (rank : 146) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60631, Q61240 | Gene names | Grb2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2). | |||||
|
CRK_HUMAN
|
||||||
| NC score | 0.053408 (rank : 147) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46108 | Gene names | CRK | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
KPCB_MOUSE
|
||||||
| NC score | 0.053154 (rank : 148) | θ value | 0.21417 (rank : 51) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P68404, P04410, P04411 | Gene names | Prkcb1, Pkcb, Prkcb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
HSH2D_MOUSE
|
||||||
| NC score | 0.053083 (rank : 149) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6VYH9, Q52KL3 | Gene names | Hsh2d, Alx | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematopoietic SH2 domain-containing protein (Hematopoietic SH2 protein) (Adaptor in lymphocytes of unknown function X). | |||||
|
KPCA_HUMAN
|
||||||
| NC score | 0.052609 (rank : 150) | θ value | 0.0736092 (rank : 44) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCB_HUMAN
|
||||||
| NC score | 0.052485 (rank : 151) | θ value | 0.21417 (rank : 50) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P05771, O43744, P05127, Q15138, Q93060, Q9UE49, Q9UE50, Q9UEH8, Q9UJ30, Q9UJ33 | Gene names | PRKCB1, PKCB, PRKCB | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C beta type (EC 2.7.11.13) (PKC-beta) (PKC-B). | |||||
|
GRB2_HUMAN
|
||||||
| NC score | 0.052410 (rank : 152) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62993, P29354, Q14450, Q63057, Q63059 | Gene names | GRB2, ASH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth factor receptor-bound protein 2 (Adapter protein GRB2) (SH2/SH3 adapter GRB2) (Protein Ash). | |||||
|
CRK_MOUSE
|
||||||
| NC score | 0.052240 (rank : 153) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64010 | Gene names | Crk, Crko | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene C-crk (P38) (Adapter molecule crk). | |||||
|
GRAP_HUMAN
|
||||||
| NC score | 0.052060 (rank : 154) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13588 | Gene names | GRAP | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-related adapter protein. | |||||
|
VAV2_HUMAN
|
||||||
| NC score | 0.052022 (rank : 155) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P52735 | Gene names | VAV2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein vav-2. | |||||
|
PA24E_HUMAN
|
||||||
| NC score | 0.051877 (rank : 156) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q3MJ16, Q6ZSC0 | Gene names | PLA2G4E | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 epsilon (EC 3.1.1.4) (cPLA2-epsilon) (Phospholipase A2 group IVE). | |||||
|
P85B_MOUSE
|
||||||
| NC score | 0.051372 (rank : 157) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O08908 | Gene names | Pik3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit beta (PI3-kinase p85- subunit beta) (PtdIns-3-kinase p85-beta). | |||||
|
RIMS3_HUMAN
|
||||||
| NC score | 0.051345 (rank : 158) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UJD0, Q92511 | Gene names | RIMS3, KIAA0237 | |||
|
Domain Architecture |

|
|||||
| Description | Regulating synaptic membrane exocytosis protein 3 (Nim3) (Rab-3- interacting molecule 3) (RIM 3) (RIM3 gamma). | |||||
|
SHE_MOUSE
|
||||||
| NC score | 0.051034 (rank : 159) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BSD5, Q3TZT0 | Gene names | She | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
VAV_MOUSE
|
||||||
| NC score | 0.050764 (rank : 160) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27870, Q8BTV7 | Gene names | Vav1, Vav | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav (p95vav). | |||||
|
SHE_HUMAN
|
||||||
| NC score | 0.050540 (rank : 161) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VZ18, Q8TEQ5 | Gene names | SHE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein E. | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.050361 (rank : 162) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
VAV_HUMAN
|
||||||
| NC score | 0.050242 (rank : 163) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15498, Q15860 | Gene names | VAV1, VAV | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene vav. | |||||
|
CAB45_MOUSE
|
||||||
| NC score | 0.049706 (rank : 164) | θ value | 0.279714 (rank : 54) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61112, Q61113 | Gene names | Sdf4, Cab45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
CU013_HUMAN
|
||||||
| NC score | 0.045270 (rank : 165) | θ value | 0.47712 (rank : 59) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95447 | Gene names | C21orf13 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C21orf13. | |||||
|
CAB45_HUMAN
|
||||||
| NC score | 0.040561 (rank : 166) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRK5, Q8NBQ3, Q9NZP7 | Gene names | SDF4, CAB45 | |||
|
Domain Architecture |

|
|||||
| Description | 45 kDa calcium-binding protein precursor (Cab45) (Stromal cell-derived factor 4) (SDF-4). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.037782 (rank : 167) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
CALB2_HUMAN
|
||||||
| NC score | 0.034771 (rank : 168) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22676, Q53HD2, Q96BK4 | Gene names | CALB2, CAB29 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR) (29 kDa calbindin). | |||||
|
CALB2_MOUSE
|
||||||
| NC score | 0.031741 (rank : 169) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08331, Q60964, Q9JM81 | Gene names | Calb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calretinin (CR). | |||||
|
MCM8_HUMAN
|
||||||
| NC score | 0.031181 (rank : 170) | θ value | 0.279714 (rank : 56) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UJA3, Q495R4, Q495R7, Q86US4, Q969I5 | Gene names | MCM8, C20orf154 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
CYLD_MOUSE
|
||||||
| NC score | 0.028344 (rank : 171) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TQ2, Q80VB3, Q8BXZ3, Q8BYL9, Q8CGB0 | Gene names | Cyld, Cyld1, Kiaa0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
CYLD_HUMAN
|
||||||
| NC score | 0.027177 (rank : 172) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQC7, O94934, Q7L3N6, Q96EH0, Q9NZX9 | Gene names | CYLD, CYLD1, KIAA0849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase CYLD (EC 3.1.2.15) (Ubiquitin thioesterase CYLD) (Ubiquitin-specific-processing protease CYLD) (Deubiquitinating enzyme CYLD). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.025827 (rank : 173) | θ value | 0.47712 (rank : 60) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
PLSI_HUMAN
|
||||||
| NC score | 0.020308 (rank : 174) | θ value | 1.38821 (rank : 66) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14651 | Gene names | PLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Plastin-1 (I-plastin) (Intestine-specific plastin). | |||||
|
MCM8_MOUSE
|
||||||
| NC score | 0.019431 (rank : 175) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWV1, Q80US2, Q80VI0 | Gene names | Mcm8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication licensing factor MCM8 (Minichromosome maintenance 8). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.018718 (rank : 176) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
TEC_HUMAN
|
||||||
| NC score | 0.016874 (rank : 177) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P42680 | Gene names | TEC, PSCTK4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
TF2LY_HUMAN
|
||||||
| NC score | 0.016639 (rank : 178) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IUE0 | Gene names | TGIF2LY, TGIFLY | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein TGIF2LY (TGFB-induced factor 2-like protein, Y- linked) (TGF(beta)induced transcription factor 2-like protein) (TGIF- like on the Y). | |||||
|
TEC_MOUSE
|
||||||
| NC score | 0.016382 (rank : 179) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 1006 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P24604, Q9R1M9, Q9WVN0, Q9WVN1, Q9WVN2, Q9WVN3 | Gene names | Tec | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase Tec (EC 2.7.10.2). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.016089 (rank : 180) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.013567 (rank : 181) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
NSF1C_MOUSE
|
||||||
| NC score | 0.012543 (rank : 182) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZ44, Q80Y15, Q8BYL3 | Gene names | Nsfl1c | |||
|
Domain Architecture |

|
|||||
| Description | NSFL1 cofactor p47 (p97 cofactor p47). | |||||
|
EFHC1_HUMAN
|
||||||
| NC score | 0.012279 (rank : 183) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JVL4, Q5XKM4, Q6E1U7, Q6E1U8, Q8WUL2, Q9NVW6 | Gene names | EFHC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing protein 1. | |||||
|
LX12E_MOUSE
|
||||||
| NC score | 0.005980 (rank : 184) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P55249, Q91YW6 | Gene names | Alox12e, Alox12-ps2, Aloxe | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 12-lipoxygenase, epidermal-type (EC 1.13.11.31) (12-LOX). | |||||
|
TGFB2_MOUSE
|
||||||
| NC score | 0.003354 (rank : 185) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 93 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27090 | Gene names | Tgfb2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming growth factor beta-2 precursor (TGF-beta-2). | |||||