Please be patient as the page loads
|
ATM_HUMAN
|
||||||
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ATM_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATM_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990469 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATR_MOUSE
|
||||||
| θ value | 1.18549e-84 (rank : 3) | NC score | 0.806565 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATR_HUMAN
|
||||||
| θ value | 1.7238e-59 (rank : 4) | NC score | 0.794003 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
FRAP_HUMAN
|
||||||
| θ value | 1.32601e-43 (rank : 5) | NC score | 0.732260 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_MOUSE
|
||||||
| θ value | 1.32601e-43 (rank : 6) | NC score | 0.732867 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
SMG1_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 7) | NC score | 0.689247 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
SMG1_MOUSE
|
||||||
| θ value | 3.85808e-43 (rank : 8) | NC score | 0.694780 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
PRKDC_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 9) | NC score | 0.666895 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| θ value | 4.42448e-31 (rank : 10) | NC score | 0.660001 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
PK3C3_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 11) | NC score | 0.474439 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3C3_HUMAN
|
||||||
| θ value | 2.0648e-12 (rank : 12) | NC score | 0.474562 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3CB_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 13) | NC score | 0.379977 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 14) | NC score | 0.379506 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CD_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 15) | NC score | 0.365132 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 16) | NC score | 0.365909 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CG_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 17) | NC score | 0.359790 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 18) | NC score | 0.357971 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
TRRAP_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 19) | NC score | 0.337551 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
TRRAP_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 20) | NC score | 0.341426 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
PI4KB_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 21) | NC score | 0.372818 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
P3C2G_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 22) | NC score | 0.317618 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PI4KB_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 23) | NC score | 0.369049 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PK3CA_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 24) | NC score | 0.325151 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 25) | NC score | 0.324958 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 26) | NC score | 0.307354 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PI4KA_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 27) | NC score | 0.312126 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 28) | NC score | 0.023056 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CJ118_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 29) | NC score | 0.027254 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
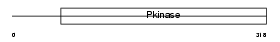
ST17A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 30) | NC score | 0.004471 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UEE5, Q8IVC8 | Gene names | STK17A, DRAK1 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase 17A (EC 2.7.11.1) (DAP kinase-related apoptosis-inducing protein kinase 1). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.026224 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SMC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.026391 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.025008 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
SYMPK_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.039717 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.004638 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.026600 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
P3C2A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.248711 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.250589 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.232768 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
SYMPK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.037723 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
NUF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.023062 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.015238 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.013464 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.012914 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
MYO1A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.008429 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBC5, Q9UQD7 | Gene names | MYO1A, MYHL | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ia (Brush border myosin I) (BBM-I) (BBMI) (Myosin I heavy chain) (MIHC). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.005943 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.015296 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
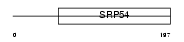
ORC4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.022428 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88708, Q9QYX1 | Gene names | Orc4l, Orc4 | |||
|
Domain Architecture |
|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
PAG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.038428 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWQ8, Q2M1Z9, Q5BKU4, Q9NYK0 | Gene names | PAG1, CBP, PAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (Transmembrane adapter protein PAG) (Csk-binding protein) (Transmembrane phosphoprotein Cbp). | |||||
|
ACOX1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.012814 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
CE152_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.017035 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
VPS18_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.032501 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
ERR2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.003503 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61539, O09067, O09146 | Gene names | Esrrb, Err-2, Err2, Nr3b2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2). | |||||
|
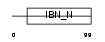
IPO8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.011066 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TMY7, Q811I3, Q8C8A9, Q8R2P6, Q8R3V7 | Gene names | Ipo8 | |||
|
Domain Architecture |
|
|||||
| Description | Importin-8 (Imp8) (Ran-binding protein 8) (RanBP8). | |||||
|
NIM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.003254 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHI9, Q8BXQ9 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
SYGP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.016294 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
FACD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.013055 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXW9, Q2LA86, Q69YP9, Q6PJN7, Q9BQ06, Q9H9T9 | Gene names | FANCD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein (Protein FACD2). | |||||
|
ROBO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.001259 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
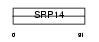
SRP14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.022864 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16254, Q3TIK2 | Gene names | Srp14 | |||
|
Domain Architecture |
|
|||||
| Description | Signal recognition particle 14 kDa protein (SRP14). | |||||
|
ZN517_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | -0.002571 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZMY9 | Gene names | ZNF517 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 517. | |||||
|
IF_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.011513 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52787 | Gene names | Gif | |||
|
Domain Architecture |

|
|||||
| Description | Gastric intrinsic factor precursor (Intrinsic factor) (IF) (INF). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.011115 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.013885 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.005077 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
RT31_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.014438 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61733, Q3TSP5, Q8VEH4, Q9D045 | Gene names | Mrps31, Imogn38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S31, mitochondrial precursor (S31mt) (MRP-S31) (Imogen 38). | |||||
|
VPS18_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.025595 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
CDKN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.016465 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16667, Q99585, Q9BPW7, Q9BY36, Q9C042, Q9C047, Q9C049, Q9C051, Q9C053 | Gene names | CDKN3, CDI1, CIP2, KAP | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 3 (EC 3.1.3.48) (EC 3.1.3.16) (CDK2- associated dual-specificity phosphatase) (Kinase-associated phosphatase) (Cyclin-dependent kinase-interacting protein 2) (Cyclin- dependent kinase interactor 1). | |||||
|
DOCK9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.005612 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.014700 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
HELC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.007852 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
KLH23_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.001419 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBE8, Q8N9B9, Q96FT8 | Gene names | KLHL23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 23. | |||||
|
NIM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.002407 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IY84 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
PLXA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.003198 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.013454 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.009928 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.009905 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.026684 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.019002 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
ATM_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q13315, O15429, Q12758, Q16551, Q93007, Q9NP02, Q9UCX7 | Gene names | ATM | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated) (A-T, mutated). | |||||
|
ATM_MOUSE
|
||||||
| NC score | 0.990469 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q62388 | Gene names | Atm | |||
|
Domain Architecture |

|
|||||
| Description | Serine-protein kinase ATM (EC 2.7.11.1) (Ataxia telangiectasia mutated homolog) (A-T, mutated homolog). | |||||
|
ATR_MOUSE
|
||||||
| NC score | 0.806565 (rank : 3) | θ value | 1.18549e-84 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9JKK8 | Gene names | Atr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein). | |||||
|
ATR_HUMAN
|
||||||
| NC score | 0.794003 (rank : 4) | θ value | 1.7238e-59 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13535, Q7KYL3, Q93051, Q9BXK4 | Gene names | ATR, FRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ATR (EC 2.7.11.1) (Ataxia telangiectasia and Rad3-related protein) (FRAP-related protein 1). | |||||
|
FRAP_MOUSE
|
||||||
| NC score | 0.732867 (rank : 5) | θ value | 1.32601e-43 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9JLN9, Q811J5, Q9CST1 | Gene names | Frap1, Frap | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
FRAP_HUMAN
|
||||||
| NC score | 0.732260 (rank : 6) | θ value | 1.32601e-43 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P42345, Q6LE87, Q96QG3, Q9Y4I3 | Gene names | FRAP1, FRAP, FRAP2 | |||
|
Domain Architecture |

|
|||||
| Description | FKBP12-rapamycin complex-associated protein (FK506-binding protein 12- rapamycin complex-associated protein 1) (Rapamycin target protein) (RAPT1) (Mammalian target of rapamycin) (mTOR). | |||||
|
SMG1_MOUSE
|
||||||
| NC score | 0.694780 (rank : 7) | θ value | 3.85808e-43 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BKX6, Q5DU42, Q6ZQC0, Q8BLU4, Q8BWJ5, Q8BXD3 | Gene names | Smg1, Kiaa0421 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1). | |||||
|
SMG1_HUMAN
|
||||||
| NC score | 0.689247 (rank : 8) | θ value | 3.85808e-43 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96Q15, O43305, Q13284, Q8NFX2, Q96QV0, Q96RW3 | Gene names | SMG1, ATX, KIAA0421, LIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SMG1 (EC 2.7.11.1) (SMG-1) (hSMG-1) (Lambda/iota protein kinase C-interacting protein) (Lambda-interacting protein) (61E3.4). | |||||
|
PRKDC_MOUSE
|
||||||
| NC score | 0.666895 (rank : 9) | θ value | 6.17384e-33 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P97313, O88187, P97928, Q307W9, Q3V2W8, Q8C2A7, Q9Z341 | Gene names | Prkdc, Xrcc7 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (P460). | |||||
|
PRKDC_HUMAN
|
||||||
| NC score | 0.660001 (rank : 10) | θ value | 4.42448e-31 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78527, P78528, Q13327, Q13337, Q14175, Q59H99, Q7Z611, Q96SE6, Q9UME3 | Gene names | PRKDC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-dependent protein kinase catalytic subunit (EC 2.7.11.1) (DNA-PK catalytic subunit) (DNA-PKcs) (DNPK1) (p460). | |||||
|
PK3C3_HUMAN
|
||||||
| NC score | 0.474562 (rank : 11) | θ value | 2.0648e-12 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8NEB9, Q15134 | Gene names | PIK3C3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3) (Phosphatidylinositol 3-kinase p100 subunit). | |||||
|
PK3C3_MOUSE
|
||||||
| NC score | 0.474439 (rank : 12) | θ value | 1.2105e-12 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6PF93, Q8R3S8 | Gene names | Pik3c3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol 3-kinase catalytic subunit type 3 (EC 2.7.1.137) (PtdIns-3-kinase type 3) (PI3-kinase type 3) (PI3K type 3) (Phosphoinositide-3-kinase class 3). | |||||
|
PK3CB_HUMAN
|
||||||
| NC score | 0.379977 (rank : 13) | θ value | 1.80886e-08 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P42338 | Gene names | PIK3CB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PK3CB_MOUSE
|
||||||
| NC score | 0.379506 (rank : 14) | θ value | 2.36244e-08 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BTI9 | Gene names | Pik3cb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit beta) (PtdIns-3-kinase p110) (PI3K) (PI3Kbeta). | |||||
|
PI4KB_HUMAN
|
||||||
| NC score | 0.372818 (rank : 15) | θ value | 2.44474e-05 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UBF8, O15096, P78405, Q9BWR6 | Gene names | PIK4CB, PI4KB | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92). | |||||
|
PI4KB_MOUSE
|
||||||
| NC score | 0.369049 (rank : 16) | θ value | 4.1701e-05 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BKC8, Q8C146 | Gene names | Pik4cb | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase beta (EC 2.7.1.67) (PtdIns 4-kinase beta) (PI4Kbeta) (PI4K-beta). | |||||
|
PK3CD_MOUSE
|
||||||
| NC score | 0.365909 (rank : 17) | θ value | 9.92553e-07 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35904 | Gene names | Pik3cd | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CD_HUMAN
|
||||||
| NC score | 0.365132 (rank : 18) | θ value | 9.92553e-07 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00329, O15445 | Gene names | PIK3CD | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta isoform (EC 2.7.1.153) (PI3-kinase p110 subunit delta) (PtdIns-3- kinase p110) (PI3K) (p110delta). | |||||
|
PK3CG_HUMAN
|
||||||
| NC score | 0.359790 (rank : 19) | θ value | 2.88788e-06 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P48736, Q8IV23, Q9BZC8 | Gene names | PIK3CG | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
PK3CG_MOUSE
|
||||||
| NC score | 0.357971 (rank : 20) | θ value | 3.77169e-06 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JHG7 | Gene names | Pik3cg, Pi3kg1 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform (EC 2.7.1.153) (PI3-kinase p110 subunit gamma) (PtdIns-3- kinase subunit p110) (PI3K) (PI3Kgamma) (p120-PI3K). | |||||
|
TRRAP_MOUSE
|
||||||
| NC score | 0.341426 (rank : 21) | θ value | 1.09739e-05 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80YV3, Q8C0Z5, Q8K104 | Gene names | Trrap | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (Tra1 homolog). | |||||
|
TRRAP_HUMAN
|
||||||
| NC score | 0.337551 (rank : 22) | θ value | 3.77169e-06 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y4A5, O75218, Q9Y631, Q9Y6H4 | Gene names | TRRAP, PAF400 | |||
|
Domain Architecture |

|
|||||
| Description | Transformation/transcription domain-associated protein (350/400 kDa PCAF-associated factor) (PAF350/400) (STAF40) (Tra1 homolog). | |||||
|
PK3CA_HUMAN
|
||||||
| NC score | 0.325151 (rank : 23) | θ value | 0.000158464 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42336, Q99762 | Gene names | PIK3CA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
PK3CA_MOUSE
|
||||||
| NC score | 0.324958 (rank : 24) | θ value | 0.000158464 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42337 | Gene names | Pik3ca | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform (EC 2.7.1.153) (PI3-kinase p110 subunit alpha) (PtdIns-3- kinase p110) (PI3K). | |||||
|
P3C2G_MOUSE
|
||||||
| NC score | 0.317618 (rank : 25) | θ value | 4.1701e-05 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O70167, O70168 | Gene names | Pik3c2g | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PI4KA_HUMAN
|
||||||
| NC score | 0.312126 (rank : 26) | θ value | 0.0252991 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P42356, Q7Z625, Q9UPG2 | Gene names | PIK4CA, PIK4 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4-kinase alpha (EC 2.7.1.67) (PI4-kinase alpha) (PtdIns-4-kinase alpha) (PI4K-alpha). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.307354 (rank : 27) | θ value | 0.00020696 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
P3C2A_MOUSE
|
||||||
| NC score | 0.250589 (rank : 28) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61194, Q61182 | Gene names | Pik3c2a, Cpk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha) (Cpk-m) (p170). | |||||
|
P3C2A_HUMAN
|
||||||
| NC score | 0.248711 (rank : 29) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00443 | Gene names | PIK3C2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-alpha) (PtdIns-3-kinase C2 alpha) (PI3K-C2alpha). | |||||
|
P3C2B_HUMAN
|
||||||
| NC score | 0.232768 (rank : 30) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00750, O95666, Q5SW99 | Gene names | PIK3C2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing beta polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-beta) (PtdIns-3-kinase C2 beta) (PI3K-C2beta) (C2-PI3K). | |||||
|
SYMPK_MOUSE
|
||||||
| NC score | 0.039717 (rank : 31) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X82 | Gene names | Sympk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
PAG1_HUMAN
|
||||||
| NC score | 0.038428 (rank : 32) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NWQ8, Q2M1Z9, Q5BKU4, Q9NYK0 | Gene names | PAG1, CBP, PAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 (Transmembrane adapter protein PAG) (Csk-binding protein) (Transmembrane phosphoprotein Cbp). | |||||
|
SYMPK_HUMAN
|
||||||
| NC score | 0.037723 (rank : 33) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92797, O00521, O00689, O00733, Q59GT5, Q8N2U5 | Gene names | SYMPK, SPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Symplekin. | |||||
|
VPS18_MOUSE
|
||||||
| NC score | 0.032501 (rank : 34) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R307, Q8BGV6, Q8BZX6, Q8C126 | Gene names | Vps18 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.027254 (rank : 35) | θ value | 0.0563607 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.026684 (rank : 36) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.026600 (rank : 37) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.026391 (rank : 38) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.026224 (rank : 39) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
VPS18_HUMAN
|
||||||
| NC score | 0.025595 (rank : 40) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P253, Q8TCG0, Q96DI3, Q9H268 | Gene names | VPS18, KIAA1475 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 18 (hVPS18). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.025008 (rank : 41) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
NUF2_MOUSE
|
||||||
| NC score | 0.023062 (rank : 42) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.023056 (rank : 43) | θ value | 0.0563607 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
SRP14_MOUSE
|
||||||
| NC score | 0.022864 (rank : 44) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P16254, Q3TIK2 | Gene names | Srp14 | |||
|
Domain Architecture |

|
|||||
| Description | Signal recognition particle 14 kDa protein (SRP14). | |||||
|
ORC4_MOUSE
|
||||||
| NC score | 0.022428 (rank : 45) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88708, Q9QYX1 | Gene names | Orc4l, Orc4 | |||
|
Domain Architecture |

|
|||||
| Description | Origin recognition complex subunit 4. | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.019002 (rank : 46) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.017035 (rank : 47) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CDKN3_HUMAN
|
||||||
| NC score | 0.016465 (rank : 48) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q16667, Q99585, Q9BPW7, Q9BY36, Q9C042, Q9C047, Q9C049, Q9C051, Q9C053 | Gene names | CDKN3, CDI1, CIP2, KAP | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 3 (EC 3.1.3.48) (EC 3.1.3.16) (CDK2- associated dual-specificity phosphatase) (Kinase-associated phosphatase) (Cyclin-dependent kinase-interacting protein 2) (Cyclin- dependent kinase interactor 1). | |||||
|
SYGP1_HUMAN
|
||||||
| NC score | 0.016294 (rank : 49) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96PV0, Q8TCS2, Q9UGE2 | Gene names | SYNGAP1, KIAA1938 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein SynGAP (Synaptic Ras-GTPase-activating protein 1) (Synaptic Ras-GAP 1) (Neuronal RasGAP). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.015296 (rank : 50) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.015238 (rank : 51) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.014700 (rank : 52) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RT31_MOUSE
|
||||||
| NC score | 0.014438 (rank : 53) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61733, Q3TSP5, Q8VEH4, Q9D045 | Gene names | Mrps31, Imogn38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28S ribosomal protein S31, mitochondrial precursor (S31mt) (MRP-S31) (Imogen 38). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.013885 (rank : 54) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.013464 (rank : 55) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.013454 (rank : 56) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
FACD2_HUMAN
|
||||||
| NC score | 0.013055 (rank : 57) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXW9, Q2LA86, Q69YP9, Q6PJN7, Q9BQ06, Q9H9T9 | Gene names | FANCD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fanconi anemia group D2 protein (Protein FACD2). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.012914 (rank : 58) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
ACOX1_MOUSE
|
||||||
| NC score | 0.012814 (rank : 59) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0H0, O35616, Q3TDG0 | Gene names | Acox1, Acox, Paox | |||
|
Domain Architecture |

|
|||||
| Description | Acyl-coenzyme A oxidase 1, peroxisomal (EC 1.3.3.6) (Palmitoyl-CoA oxidase) (AOX). | |||||
|
IF_MOUSE
|
||||||
| NC score | 0.011513 (rank : 60) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P52787 | Gene names | Gif | |||
|
Domain Architecture |

|
|||||
| Description | Gastric intrinsic factor precursor (Intrinsic factor) (IF) (INF). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.011115 (rank : 61) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
IPO8_MOUSE
|
||||||
| NC score | 0.011066 (rank : 62) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TMY7, Q811I3, Q8C8A9, Q8R2P6, Q8R3V7 | Gene names | Ipo8 | |||
|
Domain Architecture |

|
|||||
| Description | Importin-8 (Imp8) (Ran-binding protein 8) (RanBP8). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.009928 (rank : 63) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.009905 (rank : 64) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
MYO1A_HUMAN
|
||||||
| NC score | 0.008429 (rank : 65) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBC5, Q9UQD7 | Gene names | MYO1A, MYHL | |||
|
Domain Architecture |

|
|||||
| Description | Myosin Ia (Brush border myosin I) (BBM-I) (BBMI) (Myosin I heavy chain) (MIHC). | |||||
|
HELC1_HUMAN
|
||||||
| NC score | 0.007852 (rank : 66) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N3C0, O43738, Q9H1I9, Q9NTR0 | Gene names | ASCC3, HELIC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Activating signal cointegrator 1 complex subunit 3 (EC 3.6.1.-) (ASC-1 complex subunit p200) (Trip4 complex subunit p200) (Helicase, ATP binding 1). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.005943 (rank : 67) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
DOCK9_MOUSE
|
||||||
| NC score | 0.005612 (rank : 68) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BIK4, Q921Y6 | Gene names | Dock9, D14Wsu89e, Kiaa1058 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 9 (Cdc42 guanine nucleotide exchange factor zizimin 1). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.005077 (rank : 69) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.004638 (rank : 70) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
ST17A_HUMAN
|
||||||
| NC score | 0.004471 (rank : 71) | θ value | 0.125558 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UEE5, Q8IVC8 | Gene names | STK17A, DRAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 17A (EC 2.7.11.1) (DAP kinase-related apoptosis-inducing protein kinase 1). | |||||
|
ERR2_MOUSE
|
||||||
| NC score | 0.003503 (rank : 72) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61539, O09067, O09146 | Gene names | Esrrb, Err-2, Err2, Nr3b2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2). | |||||
|
NIM1_MOUSE
|
||||||
| NC score | 0.003254 (rank : 73) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BHI9, Q8BXQ9 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
PLXA3_HUMAN
|
||||||
| NC score | 0.003198 (rank : 74) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P51805 | Gene names | PLXNA3, PLXN4, SEX | |||
|
Domain Architecture |

|
|||||
| Description | Plexin-A3 precursor (Plexin-4) (Semaphorin receptor SEX). | |||||
|
NIM1_HUMAN
|
||||||
| NC score | 0.002407 (rank : 75) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IY84 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
KLH23_HUMAN
|
||||||
| NC score | 0.001419 (rank : 76) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NBE8, Q8N9B9, Q96FT8 | Gene names | KLHL23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 23. | |||||
|
ROBO1_MOUSE
|
||||||
| NC score | 0.001259 (rank : 77) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
ZN517_HUMAN
|
||||||
| NC score | -0.002571 (rank : 78) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZMY9 | Gene names | ZNF517 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 517. | |||||