Please be patient as the page loads
|
K1333_HUMAN
|
||||||
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K1333_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.990227 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
PHF7_HUMAN
|
||||||
| θ value | 1.40689e-69 (rank : 3) | NC score | 0.883734 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | 6.98233e-69 (rank : 4) | NC score | 0.891860 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 5) | NC score | 0.636086 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF6_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 6) | NC score | 0.638700 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF11_HUMAN
|
||||||
| θ value | 1.47974e-10 (rank : 7) | NC score | 0.618959 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 8) | NC score | 0.304762 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 9) | NC score | 0.308422 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 10) | NC score | 0.236043 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 11) | NC score | 0.248773 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 12) | NC score | 0.212976 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 13) | NC score | 0.337251 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 14) | NC score | 0.317584 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 15) | NC score | 0.328363 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 16) | NC score | 0.321988 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
NEDD4_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 17) | NC score | 0.062952 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
K0317_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 18) | NC score | 0.097689 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 19) | NC score | 0.097766 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
JADE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.157602 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.157301 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
ZN662_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.003951 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZS27, Q6ZNF8, Q6ZQW8 | Gene names | ZNF662 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 662. | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.045949 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.104138 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.131334 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.016141 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
UBE3C_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.068962 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
UBE3C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.064327 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
HECD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.041531 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
ZN441_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.001002 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Z8 | Gene names | ZNF441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 441. | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.134273 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
TSP3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.011034 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05895, Q6LCE0 | Gene names | Thbs3, Tsp3 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-3 precursor. | |||||
|
ATRN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.020086 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.101536 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
KCMA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.014834 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
RHPN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.014160 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWR8, Q9DBN2 | Gene names | Rhpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhophilin-2 (GTP-Rho-binding protein 2). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.105680 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.115292 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.078553 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.074643 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
ZN599_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.000412 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NL3, Q569K0, Q5PRG1 | Gene names | ZNF599 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 599. | |||||
|
ACAD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.007522 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
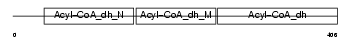
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.131983 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.059753 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
RAD9B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.013799 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6WBX7 | Gene names | Rad9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9B homolog (RAD9 homolog B) (mRAD9B). | |||||
|
TSP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.010557 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49746, Q8WV34 | Gene names | THBS3, TSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-3 precursor. | |||||
|
ATRN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.020333 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.063946 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.065969 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
TRI55_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.003941 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |
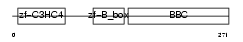
|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
AF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.062633 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.062412 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.067394 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.056257 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.053713 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.064635 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.064575 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.065162 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.064656 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.064464 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.064558 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.053836 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
MTF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050242 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
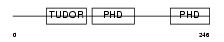
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.055514 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.054762 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.086660 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.089220 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.056930 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.059953 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.071000 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.070333 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.078154 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.079807 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.052808 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.052168 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.093135 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.051857 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.073285 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.074389 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.070911 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056133 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055190 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.058424 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.056932 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.990227 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.891860 (rank : 3) | θ value | 6.98233e-69 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.883734 (rank : 4) | θ value | 1.40689e-69 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.638700 (rank : 5) | θ value | 8.95645e-16 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.636086 (rank : 6) | θ value | 6.85773e-16 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.618959 (rank : 7) | θ value | 1.47974e-10 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.337251 (rank : 8) | θ value | 9.92553e-07 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.328363 (rank : 9) | θ value | 1.43324e-05 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.321988 (rank : 10) | θ value | 0.000121331 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.317584 (rank : 11) | θ value | 6.43352e-06 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.308422 (rank : 12) | θ value | 1.53129e-07 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.304762 (rank : 13) | θ value | 2.36244e-08 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.248773 (rank : 14) | θ value | 4.45536e-07 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.236043 (rank : 15) | θ value | 1.99992e-07 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.212976 (rank : 16) | θ value | 7.59969e-07 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.157602 (rank : 17) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.157301 (rank : 18) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.134273 (rank : 19) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.131983 (rank : 20) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.131334 (rank : 21) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.115292 (rank : 22) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.105680 (rank : 23) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.104138 (rank : 24) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.101536 (rank : 25) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
K0317_MOUSE
|
||||||
| NC score | 0.097766 (rank : 26) | θ value | 0.0148317 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CHG5, Q6P9Q1, Q80YC4, Q8C5W5 | Gene names | Kiaa0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
K0317_HUMAN
|
||||||
| NC score | 0.097689 (rank : 27) | θ value | 0.0148317 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15033, Q7LDY1, Q8IYY9 | Gene names | KIAA0317 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0317. | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.093135 (rank : 28) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.089220 (rank : 29) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.086660 (rank : 30) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.079807 (rank : 31) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.078553 (rank : 32) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.078154 (rank : 33) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.074643 (rank : 34) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.074389 (rank : 35) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.073285 (rank : 36) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.071000 (rank : 37) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.070911 (rank : 38) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.070333 (rank : 39) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
UBE3C_HUMAN
|
||||||
| NC score | 0.068962 (rank : 40) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15386, Q8TC15, Q96CR4, Q9UDU3 | Gene names | UBE3C, KIAA0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.067394 (rank : 41) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.065969 (rank : 42) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.065162 (rank : 43) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.064656 (rank : 44) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.064635 (rank : 45) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.064575 (rank : 46) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.064558 (rank : 47) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.064464 (rank : 48) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
UBE3C_MOUSE
|
||||||
| NC score | 0.064327 (rank : 49) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80U95, Q8BQZ6, Q8C7W6, Q8CDJ1, Q8VDL5 | Gene names | Ube3c, Kiaa0010 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase E3C (EC 6.3.2.-). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.063946 (rank : 50) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NEDD4_HUMAN
|
||||||
| NC score | 0.062952 (rank : 51) | θ value | 0.00390308 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P46934 | Gene names | NEDD4, KIAA0093 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase NEDD4 (EC 6.3.2.-). | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.062633 (rank : 52) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.062412 (rank : 53) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.059953 (rank : 54) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.059753 (rank : 55) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.058424 (rank : 56) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.056932 (rank : 57) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.056930 (rank : 58) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.056257 (rank : 59) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.056133 (rank : 60) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.055514 (rank : 61) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.055190 (rank : 62) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.054762 (rank : 63) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.053836 (rank : 64) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.053713 (rank : 65) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.052808 (rank : 66) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.052168 (rank : 67) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.051857 (rank : 68) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.050242 (rank : 69) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.045949 (rank : 70) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
HECD2_HUMAN
|
||||||
| NC score | 0.041531 (rank : 71) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5U5R9, Q5VZ97, Q5VZ99, Q8TCP5 | Gene names | HECTD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable E3 ubiquitin-protein ligase HECTD2 (HECT domain-containing protein 2). | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.020333 (rank : 72) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.020086 (rank : 73) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.016141 (rank : 74) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
KCMA1_HUMAN
|
||||||
| NC score | 0.014834 (rank : 75) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12791, Q12886, Q12917, Q12921, Q12960, Q13150, Q96LG8, Q9UBB0, Q9UQK6 | Gene names | KCNMA1, KCNMA | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit alpha 1 (Calcium-activated potassium channel, subfamily M subunit alpha 1) (Maxi K channel) (MaxiK) (BK channel) (K(VCA)alpha) (BKCA alpha) (KCa1.1) (Slowpoke homolog) (Slo homolog) (Slo-alpha) (Slo1) (hSlo). | |||||
|
RHPN2_MOUSE
|
||||||
| NC score | 0.014160 (rank : 76) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BWR8, Q9DBN2 | Gene names | Rhpn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhophilin-2 (GTP-Rho-binding protein 2). | |||||
|
RAD9B_MOUSE
|
||||||
| NC score | 0.013799 (rank : 77) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6WBX7 | Gene names | Rad9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle checkpoint control protein RAD9B homolog (RAD9 homolog B) (mRAD9B). | |||||
|
TSP3_MOUSE
|
||||||
| NC score | 0.011034 (rank : 78) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q05895, Q6LCE0 | Gene names | Thbs3, Tsp3 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-3 precursor. | |||||
|
TSP3_HUMAN
|
||||||
| NC score | 0.010557 (rank : 79) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49746, Q8WV34 | Gene names | THBS3, TSP3 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-3 precursor. | |||||
|
ACAD8_HUMAN
|
||||||
| NC score | 0.007522 (rank : 80) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKU7, Q9BUS8 | Gene names | ACAD8, ARC42 | |||
|
Domain Architecture |
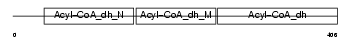
|
|||||
| Description | Acyl-CoA dehydrogenase family member 8, mitochondrial precursor (EC 1.3.99.-) (ACAD-8) (Isobutyryl-CoA dehydrogenase) (Activator- recruited cofactor 42 kDa component) (ARC42). | |||||
|
ZN662_HUMAN
|
||||||
| NC score | 0.003951 (rank : 81) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6ZS27, Q6ZNF8, Q6ZQW8 | Gene names | ZNF662 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 662. | |||||
|
TRI55_HUMAN
|
||||||
| NC score | 0.003941 (rank : 82) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYV6, Q53XX3, Q8IUD9, Q8IUE4, Q96DV2, Q96DV3, Q9BYV5 | Gene names | TRIM55, MURF2, RNF29 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 55 (Muscle-specific RING finger protein 2) (MuRF2) (MURF-2) (RING finger protein 29). | |||||
|
ZN441_HUMAN
|
||||||
| NC score | 0.001002 (rank : 83) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 754 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Z8 | Gene names | ZNF441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 441. | |||||
|
ZN599_HUMAN
|
||||||
| NC score | 0.000412 (rank : 84) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NL3, Q569K0, Q5PRG1 | Gene names | ZNF599 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 599. | |||||