Please be patient as the page loads
|
PHF7_MOUSE
|
||||||
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PHF7_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF7_HUMAN
|
||||||
| θ value | 3.37019e-164 (rank : 2) | NC score | 0.988000 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
K1333_HUMAN
|
||||||
| θ value | 6.98233e-69 (rank : 3) | NC score | 0.891860 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| θ value | 1.31681e-67 (rank : 4) | NC score | 0.891432 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 5) | NC score | 0.604817 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | 8.67504e-11 (rank : 6) | NC score | 0.600002 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF11_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 7) | NC score | 0.604029 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 8) | NC score | 0.256492 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 9) | NC score | 0.241478 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 10) | NC score | 0.305109 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 11) | NC score | 0.316413 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.299153 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.254518 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
JADE1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 14) | NC score | 0.297411 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 15) | NC score | 0.286527 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
JADE3_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 16) | NC score | 0.279927 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.250455 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 18) | NC score | 0.249524 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.280183 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 20) | NC score | 0.169005 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 21) | NC score | 0.283245 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 22) | NC score | 0.269299 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.163674 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.253716 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.187020 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.186691 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
AF10_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.224139 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.224237 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.267238 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.246962 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.175761 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.048624 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NFX1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.054493 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.173393 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.051106 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.208960 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.214398 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.218750 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.191441 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.189309 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.107480 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 0.813845 (rank : 42) | NC score | 0.224634 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 43) | NC score | 0.107723 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
DZIP3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 44) | NC score | 0.043358 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
DZIP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 45) | NC score | 0.044224 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.112064 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.117659 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.109132 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 49) | NC score | 0.046824 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
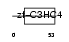
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 50) | NC score | 0.046896 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
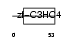
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
ZFP28_HUMAN
|
||||||
| θ value | 1.38821 (rank : 51) | NC score | 0.001935 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |
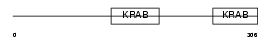
|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
LETM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.023408 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
PF21A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.105197 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
RXINP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.037613 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5U5Q9, Q60811 | Gene names | Rxrip110, Rip110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110. | |||||
|
MTF2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.101840 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
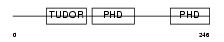
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.102880 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
ZFP28_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.001033 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10078, Q61776, Q8BT11, Q8C836 | Gene names | Zfp28, Mkr5, Zfp-28 | |||
|
Domain Architecture |
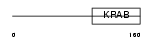
|
|||||
| Description | Zinc finger protein 28 (Zfp-28) (Protein mKR5). | |||||
|
DLL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.004035 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
ZN198_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.013378 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.017276 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
PCGF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.033540 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35226, Q16030, Q96F37 | Gene names | PCGF4, BMI1, RNF51 | |||
|
Domain Architecture |
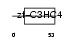
|
|||||
| Description | Polycomb group RING finger protein 4 (Polycomb complex protein BMI-1) (RING finger protein 51). | |||||
|
PCGF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.033503 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25916 | Gene names | Pcgf4, Bmi-1, Bmi1 | |||
|
Domain Architecture |
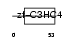
|
|||||
| Description | Polycomb group RING finger protein 4 (Polycomb complex protein BMI-1). | |||||
|
LYZL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.009746 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KX0 | Gene names | LYZL4, LYC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysozyme-like protein 4 precursor. | |||||
|
PRD15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.000622 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
TYK2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | -0.002139 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R117, O88431, Q8VE41 | Gene names | Tyk2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.079003 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.079544 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.063039 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.059248 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.064073 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.052457 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.050729 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054825 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.055384 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.082554 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.081542 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.065634 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.065595 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.066462 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.066024 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.077069 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.070673 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.070310 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.075343 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.066285 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.068814 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.069111 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053836 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.056871 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.114976 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.113830 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.061327 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.059874 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.088937 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.088663 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
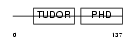
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051377 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.078670 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.077766 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.061936 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.091923 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.050589 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.078811 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.068996 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.066934 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058682 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.057732 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.062694 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.060667 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050835 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053520 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.057102 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.988000 (rank : 2) | θ value | 3.37019e-164 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.891860 (rank : 3) | θ value | 6.98233e-69 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.891432 (rank : 4) | θ value | 1.31681e-67 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.604817 (rank : 5) | θ value | 8.67504e-11 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.604029 (rank : 6) | θ value | 4.0297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.600002 (rank : 7) | θ value | 8.67504e-11 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.316413 (rank : 8) | θ value | 9.29e-05 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.305109 (rank : 9) | θ value | 5.44631e-05 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.299153 (rank : 10) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.297411 (rank : 11) | θ value | 0.00175202 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.286527 (rank : 12) | θ value | 0.00665767 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.283245 (rank : 13) | θ value | 0.0252991 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.280183 (rank : 14) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.279927 (rank : 15) | θ value | 0.00869519 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.269299 (rank : 16) | θ value | 0.0252991 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.267238 (rank : 17) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.256492 (rank : 18) | θ value | 6.43352e-06 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.254518 (rank : 19) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.253716 (rank : 20) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.250455 (rank : 21) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.249524 (rank : 22) | θ value | 0.0193708 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.246962 (rank : 23) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.241478 (rank : 24) | θ value | 2.44474e-05 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.224634 (rank : 25) | θ value | 0.813845 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.224237 (rank : 26) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.224139 (rank : 27) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.218750 (rank : 28) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.214398 (rank : 29) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.208960 (rank : 30) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.191441 (rank : 31) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.189309 (rank : 32) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.187020 (rank : 33) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.186691 (rank : 34) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.175761 (rank : 35) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.173393 (rank : 36) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.169005 (rank : 37) | θ value | 0.0252991 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.163674 (rank : 38) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.117659 (rank : 39) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.114976 (rank : 40) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.113830 (rank : 41) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.112064 (rank : 42) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.109132 (rank : 43) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.107723 (rank : 44) | θ value | 0.813845 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.107480 (rank : 45) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.105197 (rank : 46) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
MTF2_MOUSE
|
||||||
| NC score | 0.102880 (rank : 47) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.101840 (rank : 48) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.091923 (rank : 49) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.088937 (rank : 50) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.088663 (rank : 51) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.082554 (rank : 52) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.081542 (rank : 53) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.079544 (rank : 54) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.079003 (rank : 55) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.078811 (rank : 56) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.078670 (rank : 57) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.077766 (rank : 58) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.077069 (rank : 59) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.075343 (rank : 60) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.070673 (rank : 61) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.070310 (rank : 62) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JHD3D_MOUSE
|
||||||
| NC score | 0.069111 (rank : 63) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.068996 (rank : 64) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.068814 (rank : 65) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.066934 (rank : 66) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.066462 (rank : 67) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.066285 (rank : 68) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.066024 (rank : 69) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.065634 (rank : 70) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.065595 (rank : 71) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.064073 (rank : 72) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.063039 (rank : 73) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.062694 (rank : 74) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.061936 (rank : 75) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.061327 (rank : 76) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.060667 (rank : 77) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.059874 (rank : 78) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.059248 (rank : 79) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.058682 (rank : 80) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.057732 (rank : 81) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.057102 (rank : 82) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.056871 (rank : 83) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.055384 (rank : 84) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.054825 (rank : 85) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.054493 (rank : 86) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.053836 (rank : 87) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.053520 (rank : 88) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.052457 (rank : 89) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.051377 (rank : 90) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.051106 (rank : 91) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.050835 (rank : 92) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.050729 (rank : 93) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.050589 (rank : 94) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.048624 (rank : 95) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.046896 (rank : 96) | θ value | 1.38821 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.046824 (rank : 97) | θ value | 1.38821 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
DZIP3_MOUSE
|
||||||
| NC score | 0.044224 (rank : 98) | θ value | 1.06291 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7TPV2, Q80TU4, Q8BYK7 | Gene names | Dzip3, Kiaa0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3 homolog). | |||||
|
DZIP3_HUMAN
|
||||||
| NC score | 0.043358 (rank : 99) | θ value | 1.06291 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
RXINP_MOUSE
|
||||||
| NC score | 0.037613 (rank : 100) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5U5Q9, Q60811 | Gene names | Rxrip110, Rip110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoid X receptor-interacting protein 110. | |||||
|
PCGF4_HUMAN
|
||||||
| NC score | 0.033540 (rank : 101) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35226, Q16030, Q96F37 | Gene names | PCGF4, BMI1, RNF51 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 4 (Polycomb complex protein BMI-1) (RING finger protein 51). | |||||
|
PCGF4_MOUSE
|
||||||
| NC score | 0.033503 (rank : 102) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25916 | Gene names | Pcgf4, Bmi-1, Bmi1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 4 (Polycomb complex protein BMI-1). | |||||
|
LETM1_MOUSE
|
||||||
| NC score | 0.023408 (rank : 103) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2I0, Q5PQC5, Q7TMK8, Q80ZX6, Q8CGJ3, Q8K5E5 | Gene names | Letm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper-EF-hand-containing transmembrane protein 1, mitochondrial precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.017276 (rank : 104) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
ZN198_HUMAN
|
||||||
| NC score | 0.013378 (rank : 105) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBW7, O43212, O43434, O60898, Q5W0Q4, Q63HP0, Q8NE39, Q9H538, Q9UEU2 | Gene names | ZNF198, FIM, RAMP, ZMYM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 198 (Zinc finger MYM-type protein 2) (Fused in myeloproliferative disorders protein) (Rearranged in atypical myeloproliferative disorder protein). | |||||
|
LYZL4_HUMAN
|
||||||
| NC score | 0.009746 (rank : 106) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KX0 | Gene names | LYZL4, LYC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysozyme-like protein 4 precursor. | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.004035 (rank : 107) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
ZFP28_HUMAN
|
||||||
| NC score | 0.001935 (rank : 108) | θ value | 1.38821 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
ZFP28_MOUSE
|
||||||
| NC score | 0.001033 (rank : 109) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 782 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10078, Q61776, Q8BT11, Q8C836 | Gene names | Zfp28, Mkr5, Zfp-28 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 28 (Zfp-28) (Protein mKR5). | |||||
|
PRD15_HUMAN
|
||||||
| NC score | 0.000622 (rank : 110) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
TYK2_MOUSE
|
||||||
| NC score | -0.002139 (rank : 111) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R117, O88431, Q8VE41 | Gene names | Tyk2 | |||
|
Domain Architecture |

|
|||||
| Description | Non-receptor tyrosine-protein kinase TYK2 (EC 2.7.10.2). | |||||