Please be patient as the page loads
|
K0056_HUMAN
|
||||||
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0056_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
K0056_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.966023 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
CND1_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 3) | NC score | 0.320501 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2Z4, Q8R0F2, Q8R3H0, Q922B7, Q923A3, Q9CS25, Q9CT32 | Gene names | Ncapd2, Capd2, Cnap1 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (mCAP-D2) (XCAP-D2 homolog). | |||||
|
CND1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 4) | NC score | 0.316581 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 5) | NC score | 0.083664 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SAHH3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 6) | NC score | 0.071298 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FL4, Q8BIH1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.032530 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SAHH3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.068883 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96HN2, O94917 | Gene names | KIAA0828 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
PININ_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.073101 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
AP1B1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.078808 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.055724 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
AP1B1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.075088 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
ACSL6_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.035178 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WC3, Q80ZF1 | Gene names | Acsl6, Facl6 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 6 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 6) (LACS 6). | |||||
|
AP2B1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.080319 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.080321 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.032777 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.059924 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 18) | NC score | 0.052176 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
KCY_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.075574 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30085, Q53GB7, Q5SVZ0, Q96C07, Q9UBQ8, Q9UIA2 | Gene names | CMPK, CMK, UCK, UMK, UMPK | |||
|
Domain Architecture |
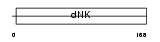
|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
KCY_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.075443 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBP5 | Gene names | Cmpk, Cmk, Uck, Umk, Umpk | |||
|
Domain Architecture |
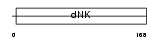
|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
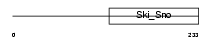
K1H8_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.018385 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76015 | Gene names | KRTHA8, HHA8, HKA8 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha8 (Hair keratin, type I Ha8). | |||||
|
KPB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.037069 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46019, Q6LAJ5, Q7Z6W0, Q96CR3, Q9UDA1 | Gene names | PHKA2, PHKLA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, liver isoform (Phosphorylase kinase alpha L subunit). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.048390 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MK07_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.008140 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.037945 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.021075 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BFSP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.023790 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
IF35_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.056737 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.024113 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.025526 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
SELV_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.053723 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
ACSL6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.030357 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKU0, O94924, O95829 | Gene names | ACSL6, ACS2, FACL6, KIAA0837, LACS5 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 6 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 6) (LACS 6). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.024023 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.022020 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.042025 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.047504 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.036032 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.037435 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.036173 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
KPB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.032557 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWJ3, Q3TN65, Q810J6 | Gene names | Phka2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, liver isoform (Phosphorylase kinase alpha L subunit). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.031121 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.017163 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.020969 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
NELFA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.033141 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.028694 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.023525 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.032527 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
BRSK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.003247 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.035877 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CUL4B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.024399 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
LIPB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.023790 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.021706 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
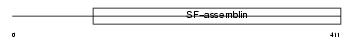
SKIL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.023090 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |
|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.033136 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
UCN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.059889 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RP3, Q9BUG0 | Gene names | UCN2, SRP, URP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Urocortin-2 precursor (Urocortin II) (Ucn II) (Stresscopin-related peptide) (Urocortin-related peptide). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.023108 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.022661 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
MCR_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.011281 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
NUAK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.002751 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||
|
AKAP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.018564 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
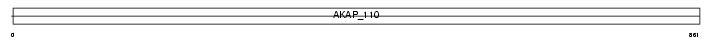
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.024957 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.019488 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.017877 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.016648 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.037090 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.039101 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.016774 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.023257 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TENS4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.025489 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.007404 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ARHGG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.009765 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
CND3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.038137 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.012220 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
MK07_MOUSE
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.005441 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.009164 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.016941 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
UBQL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.011369 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
WDR60_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.018700 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
AAK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.005578 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.018154 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.027680 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CASPA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.011828 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |
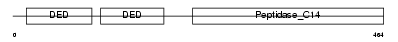
|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
CDN1C_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.022448 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
CING_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.019141 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.006981 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.018779 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ESPL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.017264 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
GBP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.014237 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
GGT4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.012676 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JP7, Q91V91 | Gene names | Ggtl3 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-glutamyltransferase 4 precursor (EC 2.3.2.2) (Gamma- glutamyltranspeptidase 4) (Gamma-glutamyltransferase-like 3) [Contains: Gamma-glutamyltransferase 4 heavy chain; Gamma- glutamyltransferase 4 light chain]. | |||||
|
HEAT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.032347 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H583, Q5T3Q8, Q9NW23 | Gene names | HEATR1, BAP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 1 (Protein BAP28). | |||||
|
NFYB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.024476 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25208, Q96IY8 | Gene names | NFYB, HAP3 | |||
|
Domain Architecture |
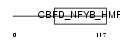
|
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
NFYB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.024451 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63139, P22569, Q3UK54 | Gene names | Nfyb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
ORCT3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.008075 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6A4L0, Q8VC89 | Gene names | Slc22a13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Organic cation transporter-like 3 (Solute carrier family 22 member 13). | |||||
|
RABE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.034602 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.016166 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.024996 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TTC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.017830 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |
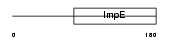
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
BFSP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.018277 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13515, Q9HBW5 | Gene names | BFSP2 | |||
|
Domain Architecture |
|
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L). | |||||
|
CT043_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.015528 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BY42, Q9BYL7, Q9HCV9, Q9NX29, Q9NZZ8, Q9P002, Q9UHW3 | Gene names | C20orf43 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf43. | |||||
|
MFRN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.006967 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0Z5, Q8K4J8, Q8K4J9, Q8R578 | Gene names | Slc25a28, Mfrn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.009824 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
NOC3L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.018744 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
PARG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.013441 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
SOX9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.009561 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.017413 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TTC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.015622 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |
|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
VPP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.007635 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920R6, Q8CJ79, Q920B5 | Gene names | Atp6v0a4, Atp6n1b | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 4 (V- ATPase 116 kDa isoform a4) (Vacuolar proton translocating ATPase 116 kDa subunit a kidney isoform). | |||||
|
RTDR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.053839 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
K0056_MOUSE
|
||||||
| NC score | 0.966023 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQK0, Q9CS21 | Gene names | Kiaa0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
CND1_MOUSE
|
||||||
| NC score | 0.320501 (rank : 3) | θ value | 3.64472e-09 (rank : 3) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2Z4, Q8R0F2, Q8R3H0, Q922B7, Q923A3, Q9CS25, Q9CT32 | Gene names | Ncapd2, Capd2, Cnap1 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (mCAP-D2) (XCAP-D2 homolog). | |||||
|
CND1_HUMAN
|
||||||
| NC score | 0.316581 (rank : 4) | θ value | 2.36244e-08 (rank : 4) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15021, Q8N6U3 | Gene names | NCAPD2, CAPD2, CNAP1, KIAA0159 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 1 (Non-SMC condensin I complex subunit D2) (Chromosome condensation-related SMC-associated protein 1) (Chromosome-associated protein D2) (hCAP-D2) (XCAP-D2 homolog). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.083664 (rank : 5) | θ value | 0.00390308 (rank : 5) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
AP2B1_MOUSE
|
||||||
| NC score | 0.080321 (rank : 6) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9DBG3, Q80XJ4 | Gene names | Ap2b1, Clapb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP2B1_HUMAN
|
||||||
| NC score | 0.080319 (rank : 7) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P63010, P21851, Q96J19 | Gene names | AP2B1, CLAPB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit beta-1 (Adapter-related protein complex 2 beta-1 subunit) (Beta-adaptin) (Plasma membrane adaptor HA2/AP2 adaptin beta subunit) (Clathrin assembly protein complex 2 beta large chain) (AP105B). | |||||
|
AP1B1_HUMAN
|
||||||
| NC score | 0.078808 (rank : 8) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q10567, P78436 | Gene names | AP1B1, ADTB1, BAM22 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
KCY_HUMAN
|
||||||
| NC score | 0.075574 (rank : 9) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30085, Q53GB7, Q5SVZ0, Q96C07, Q9UBQ8, Q9UIA2 | Gene names | CMPK, CMK, UCK, UMK, UMPK | |||
|
Domain Architecture |

|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
KCY_MOUSE
|
||||||
| NC score | 0.075443 (rank : 10) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBP5 | Gene names | Cmpk, Cmk, Uck, Umk, Umpk | |||
|
Domain Architecture |

|
|||||
| Description | UMP-CMP kinase (EC 2.7.4.14) (Cytidylate kinase) (Deoxycytidylate kinase) (Cytidine monophosphate kinase) (Uridine monophosphate/cytidine monophosphate kinase) (UMP/CMP kinase) (UMP/CMPK) (Uridine monophosphate kinase). | |||||
|
AP1B1_MOUSE
|
||||||
| NC score | 0.075088 (rank : 11) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35643, Q922E2 | Gene names | Ap1b1, Adtb1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit beta-1 (Adapter-related protein complex 1 beta-1 subunit) (Beta-adaptin 1) (Adaptor protein complex AP-1 beta-1 subunit) (Golgi adaptor HA1/AP1 adaptin beta subunit) (Clathrin assembly protein complex 1 beta large chain). | |||||
|
PININ_HUMAN
|
||||||
| NC score | 0.073101 (rank : 12) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H307, O60899, Q53EM7, Q6P5X4, Q7KYL1, Q99738, Q9UHZ9, Q9UQR9 | Gene names | PNN, DRS, MEMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin (140 kDa nuclear and cell adhesion-related phosphoprotein) (Domain-rich serine protein) (DRS-protein) (DRSP) (Melanoma metastasis clone A protein) (Desmosome-associated protein) (SR-like protein) (Nuclear protein SDK3). | |||||
|
SAHH3_MOUSE
|
||||||
| NC score | 0.071298 (rank : 13) | θ value | 0.0148317 (rank : 6) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FL4, Q8BIH1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
SAHH3_HUMAN
|
||||||
| NC score | 0.068883 (rank : 14) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96HN2, O94917 | Gene names | KIAA0828 | |||
|
Domain Architecture |

|
|||||
| Description | Putative adenosylhomocysteinase 3 (EC 3.3.1.1) (S-adenosyl-L- homocysteine hydrolase 3) (AdoHcyase 3). | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.059924 (rank : 15) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
UCN2_HUMAN
|
||||||
| NC score | 0.059889 (rank : 16) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96RP3, Q9BUG0 | Gene names | UCN2, SRP, URP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Urocortin-2 precursor (Urocortin II) (Ucn II) (Stresscopin-related peptide) (Urocortin-related peptide). | |||||
|
IF35_HUMAN
|
||||||
| NC score | 0.056737 (rank : 17) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00303, Q8N978 | Gene names | EIF3S5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.055724 (rank : 18) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
RTDR1_HUMAN
|
||||||
| NC score | 0.053839 (rank : 19) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHP6 | Gene names | RTDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhabdoid tumor deletion region protein 1. | |||||
|
SELV_HUMAN
|
||||||
| NC score | 0.053723 (rank : 20) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P59797 | Gene names | SELV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Selenoprotein V. | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.052176 (rank : 21) | θ value | 0.365318 (rank : 18) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.048390 (rank : 22) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.047504 (rank : 23) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.042025 (rank : 24) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.039101 (rank : 25) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
CND3_HUMAN
|
||||||
| NC score | 0.038137 (rank : 26) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BPX3, Q96SV9, Q9BUR3, Q9BVY1, Q9H914, Q9H9Z6, Q9HBI9 | Gene names | NCAPG, CAPG, NYMEL3 | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 3 (Non-SMC condensin I complex subunit G) (Chromosome-associated protein G) (Condensin subunit CAP-G) (hCAP-G) (XCAP-G homolog) (Antigen NY-MEL-3). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.037945 (rank : 27) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.037435 (rank : 28) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.037090 (rank : 29) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
KPB2_HUMAN
|
||||||
| NC score | 0.037069 (rank : 30) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46019, Q6LAJ5, Q7Z6W0, Q96CR3, Q9UDA1 | Gene names | PHKA2, PHKLA | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, liver isoform (Phosphorylase kinase alpha L subunit). | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.036173 (rank : 31) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.036032 (rank : 32) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.035877 (rank : 33) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ACSL6_MOUSE
|
||||||
| NC score | 0.035178 (rank : 34) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WC3, Q80ZF1 | Gene names | Acsl6, Facl6 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 6 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 6) (LACS 6). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.034602 (rank : 35) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
NELFA_MOUSE
|
||||||
| NC score | 0.033141 (rank : 36) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BG30, Q8BVE4, Q8VEI7, Q9CSJ9, Q9Z1V9 | Gene names | Whsc2, Nelfa, Whsc2h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Negative elongation factor A (NELF-A) (Wolf-Hirschhorn syndrome candidate 2 homolog) (mWHSC2). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.033136 (rank : 37) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.032777 (rank : 38) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
KPB2_MOUSE
|
||||||
| NC score | 0.032557 (rank : 39) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BWJ3, Q3TN65, Q810J6 | Gene names | Phka2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphorylase b kinase regulatory subunit alpha, liver isoform (Phosphorylase kinase alpha L subunit). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.032530 (rank : 40) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.032527 (rank : 41) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
HEAT1_HUMAN
|
||||||
| NC score | 0.032347 (rank : 42) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H583, Q5T3Q8, Q9NW23 | Gene names | HEATR1, BAP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HEAT repeat-containing protein 1 (Protein BAP28). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.031121 (rank : 43) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
ACSL6_HUMAN
|
||||||
| NC score | 0.030357 (rank : 44) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKU0, O94924, O95829 | Gene names | ACSL6, ACS2, FACL6, KIAA0837, LACS5 | |||
|
Domain Architecture |

|
|||||
| Description | Long-chain-fatty-acid--CoA ligase 6 (EC 6.2.1.3) (Long-chain acyl-CoA synthetase 6) (LACS 6). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.028694 (rank : 45) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.027680 (rank : 46) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.025526 (rank : 47) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
TENS4_HUMAN
|
||||||
| NC score | 0.025489 (rank : 48) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IZW8, Q71RB7, Q8WV64, Q96JV4 | Gene names | TNS4, CTEN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor (C-terminal tensin-like protein). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.024996 (rank : 49) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.024957 (rank : 50) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
NFYB_HUMAN
|
||||||
| NC score | 0.024476 (rank : 51) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25208, Q96IY8 | Gene names | NFYB, HAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
NFYB_MOUSE
|
||||||
| NC score | 0.024451 (rank : 52) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63139, P22569, Q3UK54 | Gene names | Nfyb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
CUL4B_HUMAN
|
||||||
| NC score | 0.024399 (rank : 53) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13620, Q6PIE4, Q6UP07, Q7Z673, Q9BY37, Q9UEB7, Q9UED7 | Gene names | CUL4B, KIAA0695 | |||
|
Domain Architecture |

|
|||||
| Description | Cullin-4B (CUL-4B). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.024113 (rank : 54) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.024023 (rank : 55) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
BFSP2_MOUSE
|
||||||
| NC score | 0.023790 (rank : 56) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NVD9, Q63832, Q6P5N4, Q8VDD6 | Gene names | Bfsp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein). | |||||
|
LIPB1_HUMAN
|
||||||
| NC score | 0.023790 (rank : 57) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.023525 (rank : 58) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.023257 (rank : 59) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.023108 (rank : 60) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
SKIL_HUMAN
|
||||||
| NC score | 0.023090 (rank : 61) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12757, O00464, P12756, Q07501 | Gene names | SKIL, SNO | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.022661 (rank : 62) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
CDN1C_HUMAN
|
||||||
| NC score | 0.022448 (rank : 63) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49918 | Gene names | CDKN1C, KIP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.022020 (rank : 64) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.021706 (rank : 65) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.021075 (rank : 66) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.020969 (rank : 67) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.019488 (rank : 68) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.019141 (rank : 69) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.018779 (rank : 70) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NOC3L_MOUSE
|
||||||
| NC score | 0.018744 (rank : 71) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VI84, Q9JKM3 | Gene names | Noc3l, Ad24, Fad24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar complex protein 3 homolog (NOC3 protein homolog) (NOC3-like protein) (Nucleolar complex-associated protein 3-like protein) (Factor for adipocyte differentiation 24). | |||||
|
WDR60_HUMAN
|
||||||
| NC score | 0.018700 (rank : 72) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WVS4, Q9NW58 | Gene names | WDR60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 60. | |||||
|
AKAP3_MOUSE
|
||||||
| NC score | 0.018564 (rank : 73) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
K1H8_HUMAN
|
||||||
| NC score | 0.018385 (rank : 74) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O76015 | Gene names | KRTHA8, HHA8, HKA8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha8 (Hair keratin, type I Ha8). | |||||
|
BFSP2_HUMAN
|
||||||
| NC score | 0.018277 (rank : 75) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13515, Q9HBW5 | Gene names | BFSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.018154 (rank : 76) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.017877 (rank : 77) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TTC4_MOUSE
|
||||||
| NC score | 0.017830 (rank : 78) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R3H9 | Gene names | Ttc4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.017413 (rank : 79) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ESPL1_MOUSE
|
||||||
| NC score | 0.017264 (rank : 80) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P60330 | Gene names | Espl1, Esp1, Kiaa0165 | |||
|
Domain Architecture |

|
|||||
| Description | Separin (EC 3.4.22.49) (Separase) (Caspase-like protein ESPL1) (Extra spindle poles-like 1 protein). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.017163 (rank : 81) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.016941 (rank : 82) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.016774 (rank : 83) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
MYC_MOUSE
|
||||||
| NC score | 0.016648 (rank : 84) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01108, P70247, Q3UM70, Q61422 | Gene names | Myc | |||
|
Domain Architecture |

|
|||||
| Description | Myc proto-oncogene protein (c-Myc) (Transcription factor p64). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.016166 (rank : 85) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
TTC4_HUMAN
|
||||||
| NC score | 0.015622 (rank : 86) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95801, Q9H3I2 | Gene names | TTC4 | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 4 (TPR repeat protein 4). | |||||
|
CT043_HUMAN
|
||||||
| NC score | 0.015528 (rank : 87) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BY42, Q9BYL7, Q9HCV9, Q9NX29, Q9NZZ8, Q9P002, Q9UHW3 | Gene names | C20orf43 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf43. | |||||
|
GBP4_HUMAN
|
||||||
| NC score | 0.014237 (rank : 88) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.013441 (rank : 89) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
GGT4_MOUSE
|
||||||
| NC score | 0.012676 (rank : 90) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JP7, Q91V91 | Gene names | Ggtl3 | |||
|
Domain Architecture |

|
|||||
| Description | Gamma-glutamyltransferase 4 precursor (EC 2.3.2.2) (Gamma- glutamyltranspeptidase 4) (Gamma-glutamyltransferase-like 3) [Contains: Gamma-glutamyltransferase 4 heavy chain; Gamma- glutamyltransferase 4 light chain]. | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.012220 (rank : 91) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
CASPA_HUMAN
|
||||||
| NC score | 0.011828 (rank : 92) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92851, Q8WYQ8, Q99845, Q9Y2U6, Q9Y2U7 | Gene names | CASP10, MCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase-10 precursor (EC 3.4.22.-) (CASP-10) (ICE-like apoptotic protease 4) (Apoptotic protease Mch-4) (FAS-associated death domain protein interleukin-1B-converting enzyme 2) (FLICE2) [Contains: Caspase-10 subunit p23/17; Caspase-10 subunit p12]. | |||||
|
UBQL2_MOUSE
|
||||||
| NC score | 0.011369 (rank : 93) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZM0, Q7TSJ8, Q8VDH9 | Gene names | Ubqln2, Plic2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.011281 (rank : 94) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.009824 (rank : 95) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
ARHGG_HUMAN
|
||||||
| NC score | 0.009765 (rank : 96) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VV41, Q86TF0, Q99434 | Gene names | ARHGEF16, NBR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
SOX9_HUMAN
|
||||||
| NC score | 0.009561 (rank : 97) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P48436 | Gene names | SOX9 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-9. | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.009164 (rank : 98) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
MK07_HUMAN
|
||||||
| NC score | 0.008140 (rank : 99) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1087 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13164, Q16634 | Gene names | MAPK7, ERK4, ERK5, PRKM7 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (ERK4) (BMK1 kinase). | |||||
|
ORCT3_MOUSE
|
||||||
| NC score | 0.008075 (rank : 100) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6A4L0, Q8VC89 | Gene names | Slc22a13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Organic cation transporter-like 3 (Solute carrier family 22 member 13). | |||||
|
VPP4_MOUSE
|
||||||
| NC score | 0.007635 (rank : 101) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920R6, Q8CJ79, Q920B5 | Gene names | Atp6v0a4, Atp6n1b | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 4 (V- ATPase 116 kDa isoform a4) (Vacuolar proton translocating ATPase 116 kDa subunit a kidney isoform). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.007404 (rank : 102) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.006981 (rank : 103) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
MFRN2_MOUSE
|
||||||
| NC score | 0.006967 (rank : 104) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0Z5, Q8K4J8, Q8K4J9, Q8R578 | Gene names | Slc25a28, Mfrn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitoferrin-2 (Mitochondrial iron transporter 2) (Solute carrier family 25 member 28) (Mitochondrial RNA-splicing protein 3/4 homolog) (MRS3/4). | |||||
|
AAK1_HUMAN
|
||||||
| NC score | 0.005578 (rank : 105) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1077 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q2M2I8, Q4ZFZ3, Q53RX6, Q9UPV4 | Gene names | AAK1, KIAA1048 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP2-associated protein kinase 1 (EC 2.7.11.1) (Adaptor-associated kinase 1). | |||||
|
MK07_MOUSE
|
||||||
| NC score | 0.005441 (rank : 106) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 1054 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVS8 | Gene names | Mapk7, Erk5 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase 7 (EC 2.7.11.24) (Extracellular signal-regulated kinase 5) (ERK-5) (BMK1 kinase). | |||||
|
BRSK2_HUMAN
|
||||||
| NC score | 0.003247 (rank : 107) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 897 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IWQ3, O60843, O95099, Q5J5B4, Q8TB60 | Gene names | BRSK2, C11orf7, PEN11B, STK29 | |||
|
Domain Architecture |

|
|||||
| Description | BR serine/threonine-protein kinase 2 (EC 2.7.11.1) (Serine/threonine- protein kinase 29) (SAD1B). | |||||
|
NUAK1_HUMAN
|
||||||
| NC score | 0.002751 (rank : 108) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 107 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60285, Q96KA8 | Gene names | NUAK1, ARK5, KIAA0537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 1 (EC 2.7.11.1) (AMPK-related protein kinase 5). | |||||