Please be patient as the page loads
|
PARG_MOUSE
|
||||||
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PARG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.966839 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PARG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 3) | NC score | 0.103855 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.048478 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 5) | NC score | 0.067294 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CDS2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.057376 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 7) | NC score | 0.029780 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 8) | NC score | 0.058623 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
M3K7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.008473 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43318, O43317, O43319, Q5TDN2, Q5TDN3 | Gene names | MAP3K7, TAK1 | |||
|
Domain Architecture |
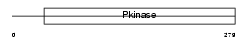
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
M3K7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.008532 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62073 | Gene names | Map3k7, Tak1 | |||
|
Domain Architecture |
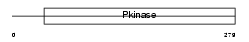
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
R51A1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.081394 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96B01, O43403, Q7Z779 | Gene names | RAD51AP1, PIR51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.061042 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
MCPH1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.040817 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.014175 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
AKAP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.057862 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75969, O75945, Q9UM61 | Gene names | AKAP3, AKAP110, SOB1 | |||
|
Domain Architecture |
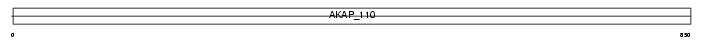
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte- binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95). | |||||
|
AKAP3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.058968 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
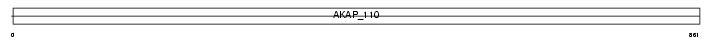
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.048321 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
NOL8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.036746 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.022568 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
RNPS1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.036365 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.036365 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.035350 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.034196 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
TOPB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.030429 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.045813 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.010829 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
ZNF23_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.000612 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||
|
ACS2L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.035508 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NB1, Q8K0M6 | Gene names | Acss1, Acas2, Acas2l | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (AceCS2) (Acyl-CoA synthetase short-chain family member 1). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.036855 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
DX26B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.040210 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
IRF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.018539 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15314 | Gene names | Irf1 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
TIG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.037415 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99969 | Gene names | RARRES2, TIG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor responder protein 2 precursor (Tazarotene- induced gene 2 protein) (RAR-responsive protein TIG2). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.009659 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.026665 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.028854 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GUC2D_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.007723 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.033881 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CDS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.035100 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95674, Q5TDY2, Q5TDY3, Q5TDY4, Q5TDY5, Q9BYK5, Q9NTT2 | Gene names | CDS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.023360 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
DPOLN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.019198 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
GALT5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.006889 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.015019 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PAF49_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.023965 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
RIMB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.013359 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U40 | Gene names | Rimbp2, Kiaa0318, Rbp2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.009020 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
ACS2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.028192 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUB1, Q5TF42, Q8IV99, Q8N234, Q96JI1, Q96JX6, Q9NU28 | Gene names | ACSS1, ACAS2L, KIAA1846 | |||
|
Domain Architecture |
|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (Acyl- CoA synthetase short-chain family member 1). | |||||
|
ALEX_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.015302 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.012632 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
CRAL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.013994 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z275 | Gene names | Rlbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular retinaldehyde-binding protein (CRALBP). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.039788 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UT14B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.012382 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.016259 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
K0056_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.013441 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
K1543_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.012817 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.966839 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.103855 (rank : 3) | θ value | 0.0252991 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
R51A1_HUMAN
|
||||||
| NC score | 0.081394 (rank : 4) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96B01, O43403, Q7Z779 | Gene names | RAD51AP1, PIR51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.067294 (rank : 5) | θ value | 0.0961366 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.061042 (rank : 6) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
AKAP3_MOUSE
|
||||||
| NC score | 0.058968 (rank : 7) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.058623 (rank : 8) | θ value | 0.813845 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AKAP3_HUMAN
|
||||||
| NC score | 0.057862 (rank : 9) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75969, O75945, Q9UM61 | Gene names | AKAP3, AKAP110, SOB1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte- binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95). | |||||
|
CDS2_MOUSE
|
||||||
| NC score | 0.057376 (rank : 10) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.048478 (rank : 11) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.048321 (rank : 12) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.045813 (rank : 13) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
MCPH1_HUMAN
|
||||||
| NC score | 0.040817 (rank : 14) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
DX26B_HUMAN
|
||||||
| NC score | 0.040210 (rank : 15) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.039788 (rank : 16) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TIG2_HUMAN
|
||||||
| NC score | 0.037415 (rank : 17) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99969 | Gene names | RARRES2, TIG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor responder protein 2 precursor (Tazarotene- induced gene 2 protein) (RAR-responsive protein TIG2). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.036855 (rank : 18) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
NOL8_HUMAN
|
||||||
| NC score | 0.036746 (rank : 19) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q76FK4, Q5TCC7, Q5TCC8, Q5TCD3, Q5TCD5, Q5TCD6, Q5TCD7, Q76D35, Q7L3E2, Q9H586, Q9H795, Q9H7W7, Q9H9J6, Q9NWA4, Q9NWM4 | Gene names | NOL8, NOP132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 8 (Nucleolar protein Nop132). | |||||
|
RNPS1_HUMAN
|
||||||
| NC score | 0.036365 (rank : 20) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15287, O75308, Q8WY42, Q9NYG3 | Gene names | RNPS1, LDC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1 (SR-related protein LDC2). | |||||
|
RNPS1_MOUSE
|
||||||
| NC score | 0.036365 (rank : 21) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99M28, Q3TMJ1, Q922H8 | Gene names | Rnps1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein with serine-rich domain 1. | |||||
|
ACS2L_MOUSE
|
||||||
| NC score | 0.035508 (rank : 22) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NB1, Q8K0M6 | Gene names | Acss1, Acas2, Acas2l | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (AceCS2) (Acyl-CoA synthetase short-chain family member 1). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.035350 (rank : 23) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
CDS2_HUMAN
|
||||||
| NC score | 0.035100 (rank : 24) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95674, Q5TDY2, Q5TDY3, Q5TDY4, Q5TDY5, Q9BYK5, Q9NTT2 | Gene names | CDS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.034196 (rank : 25) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.033881 (rank : 26) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
TOPB1_HUMAN
|
||||||
| NC score | 0.030429 (rank : 27) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.029780 (rank : 28) | θ value | 0.62314 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.028854 (rank : 29) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ACS2L_HUMAN
|
||||||
| NC score | 0.028192 (rank : 30) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUB1, Q5TF42, Q8IV99, Q8N234, Q96JI1, Q96JX6, Q9NU28 | Gene names | ACSS1, ACAS2L, KIAA1846 | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (Acyl- CoA synthetase short-chain family member 1). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.026665 (rank : 31) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PAF49_MOUSE
|
||||||
| NC score | 0.023965 (rank : 32) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q76KJ5, Q8K0Y9 | Gene names | Cd3eap, Ase1, Paf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.023360 (rank : 33) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.022568 (rank : 34) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
DPOLN_HUMAN
|
||||||
| NC score | 0.019198 (rank : 35) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z5Q5, Q4TTW4, Q6ZNF4 | Gene names | POLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase nu (EC 2.7.7.7). | |||||
|
IRF1_MOUSE
|
||||||
| NC score | 0.018539 (rank : 36) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P15314 | Gene names | Irf1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 1 (IRF-1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.016259 (rank : 37) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
ALEX_HUMAN
|
||||||
| NC score | 0.015302 (rank : 38) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P84996 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALEX (Alternative gene product encoded by XL-exon). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.015019 (rank : 39) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.014175 (rank : 40) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CRAL_MOUSE
|
||||||
| NC score | 0.013994 (rank : 41) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z275 | Gene names | Rlbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular retinaldehyde-binding protein (CRALBP). | |||||
|
K0056_HUMAN
|
||||||
| NC score | 0.013441 (rank : 42) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P42695 | Gene names | KIAA0056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0056. | |||||
|
RIMB2_MOUSE
|
||||||
| NC score | 0.013359 (rank : 43) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80U40 | Gene names | Rimbp2, Kiaa0318, Rbp2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.012817 (rank : 44) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.012632 (rank : 45) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
UT14B_MOUSE
|
||||||
| NC score | 0.012382 (rank : 46) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6EJB6, Q6EJB5, Q8BL60 | Gene names | Utp14b, Jsd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog B (Juvenile spermatogonial depletion protein). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.010829 (rank : 47) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.009659 (rank : 48) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.009020 (rank : 49) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
M3K7_MOUSE
|
||||||
| NC score | 0.008532 (rank : 50) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62073 | Gene names | Map3k7, Tak1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
M3K7_HUMAN
|
||||||
| NC score | 0.008473 (rank : 51) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43318, O43317, O43319, Q5TDN2, Q5TDN3 | Gene names | MAP3K7, TAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
GUC2D_HUMAN
|
||||||
| NC score | 0.007723 (rank : 52) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02846, Q6LEA7 | Gene names | GUCY2D, CORD6, GUC1A4, GUC2D, RETGC, RETGC1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinal guanylyl cyclase 1 precursor (EC 4.6.1.2) (Guanylate cyclase 2D, retinal) (RETGC-1) (Rod outer segment membrane guanylate cyclase) (ROS-GC). | |||||
|
GALT5_HUMAN
|
||||||
| NC score | 0.006889 (rank : 53) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z7M9, Q9UGK7, Q9UHL6 | Gene names | GALNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) (Protein-UDP acetylgalactosaminyltransferase 5) (UDP- GalNAc:polypeptide N-acetylgalactosaminyltransferase 5) (Polypeptide GalNAc transferase 5) (GalNAc-T5) (pp-GaNTase 5). | |||||
|
ZNF23_HUMAN
|
||||||
| NC score | 0.000612 (rank : 54) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17027, Q96IT3, Q9UG42 | Gene names | ZNF23, KOX16, ZNF359 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 23 (Zinc finger protein 359) (Zinc finger protein KOX16). | |||||