Please be patient as the page loads
|
PARG_HUMAN
|
||||||
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PARG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PARG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.966839 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 3) | NC score | 0.125856 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 4) | NC score | 0.111705 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 5) | NC score | 0.085572 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.110644 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.081265 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.080997 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.080130 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.115778 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.103772 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.037282 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.101847 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HSP72_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.024816 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
IF38_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.076840 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.022381 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
UBP1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.040021 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.023687 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
NAF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.027891 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.069298 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.061937 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CLUA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.049690 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AJ1, O75138, Q65ZA3, Q9H8R4, Q9H8T1 | Gene names | CLUAP1, KIAA0643 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
NUCL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.036642 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
HSP72_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.023073 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.034665 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
R51A1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.083150 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96B01, O43403, Q7Z779 | Gene names | RAD51AP1, PIR51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein). | |||||
|
TBX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.018282 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.062053 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AKAP3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.058896 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
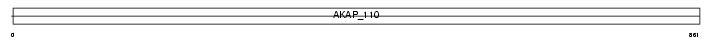
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.009857 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.049868 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
FGD6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.020139 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.048980 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.063961 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPM_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.056484 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
TRI44_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.044633 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
AKAP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.058097 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75969, O75945, Q9UM61 | Gene names | AKAP3, AKAP110, SOB1 | |||
|
Domain Architecture |
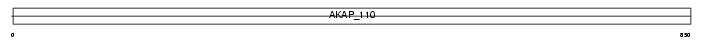
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte- binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95). | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.008514 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.043913 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
APC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.036464 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.033678 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.023960 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.058989 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.051282 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
CI039_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.020180 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
DX26B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.047436 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
LIMA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.025030 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.102416 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
STK10_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.007867 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.027306 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.067773 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TOP2A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.034386 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ACS2L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.034735 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NB1, Q8K0M6 | Gene names | Acss1, Acas2, Acas2l | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (AceCS2) (Acyl-CoA synthetase short-chain family member 1). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.052275 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
K1383_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.045989 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
MAP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.048749 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.024973 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
TOP2B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.034730 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02880, Q13600, Q9UMG8, Q9UQP8 | Gene names | TOP2B | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.058984 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.013089 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.031192 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.005273 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
HNRPC_MOUSE
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.018987 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |
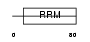
|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.013522 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
LRC27_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.014706 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YS5, Q9D6Z2 | Gene names | Lrrc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.045970 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.033570 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.016052 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.033414 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.032041 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
NPM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.036720 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.026202 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.026476 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WDFY3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.014754 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
ACS2L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.027542 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUB1, Q5TF42, Q8IV99, Q8N234, Q96JI1, Q96JX6, Q9NU28 | Gene names | ACSS1, ACAS2L, KIAA1846 | |||
|
Domain Architecture |
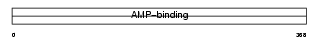
|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (Acyl- CoA synthetase short-chain family member 1). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.045605 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.028647 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CDS2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.035439 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
CF152_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.015726 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.082828 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
MASTL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.002336 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.029269 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PRGC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.016997 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RBM28_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.015997 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
SDCG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.027837 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |
|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
SNIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.030706 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BIZ6, Q3V106, Q8BIZ4 | Gene names | Snip1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
TOIP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.029729 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.034379 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
CK060_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.016211 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQC8 | Gene names | C11orf60, C11orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0360 protein C11orf60. | |||||
|
CLUA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.033114 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
DX26B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.026927 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BND4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
DZIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.011583 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.059395 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.030470 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.012600 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
WDFY3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.013948 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.016252 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.056625 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.064270 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.061199 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.066031 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.051272 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.966839 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.125856 (rank : 3) | θ value | 0.00102713 (rank : 3) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.115778 (rank : 4) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.111705 (rank : 5) | θ value | 0.00390308 (rank : 4) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.110644 (rank : 6) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.103772 (rank : 7) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.102416 (rank : 8) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.101847 (rank : 9) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.085572 (rank : 10) | θ value | 0.0113563 (rank : 5) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
R51A1_HUMAN
|
||||||
| NC score | 0.083150 (rank : 11) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96B01, O43403, Q7Z779 | Gene names | RAD51AP1, PIR51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RAD51-associated protein 1 (RAD51-interacting protein). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.082828 (rank : 12) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.081265 (rank : 13) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.080997 (rank : 14) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.080130 (rank : 15) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
IF38_HUMAN
|
||||||
| NC score | 0.076840 (rank : 16) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99613, O00215 | Gene names | EIF3S8 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 8 (eIF3 p110) (eIF3c). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.069298 (rank : 17) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.067773 (rank : 18) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.066031 (rank : 19) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.064270 (rank : 20) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.063961 (rank : 21) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.062053 (rank : 22) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.061937 (rank : 23) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.061199 (rank : 24) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.059395 (rank : 25) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.058989 (rank : 26) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.058984 (rank : 27) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AKAP3_MOUSE
|
||||||
| NC score | 0.058896 (rank : 28) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88987 | Gene names | Akap3, Akap110 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110). | |||||
|
AKAP3_HUMAN
|
||||||
| NC score | 0.058097 (rank : 29) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75969, O75945, Q9UM61 | Gene names | AKAP3, AKAP110, SOB1 | |||
|
Domain Architecture |
|
|||||
| Description | A-kinase anchor protein 3 (Protein kinase A-anchoring protein 3) (PRKA3) (A-kinase anchor protein 110 kDa) (AKAP 110) (Sperm oocyte- binding protein) (Fibrousheathin-1) (Fibrousheathin I) (Fibrous sheath protein of 95 kDa) (FSP95). | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.056625 (rank : 30) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
NPM_HUMAN
|
||||||
| NC score | 0.056484 (rank : 31) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P06748, P08693, Q12826, Q13440, Q13441, Q14115, Q5EU94, Q5EU95, Q5EU96, Q5EU97, Q5EU98, Q5EU99, Q6V962, Q8WTW5, Q96AT6, Q96DC4, Q96EA5 | Gene names | NPM1, NPM | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.052275 (rank : 32) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.051282 (rank : 33) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.051272 (rank : 34) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.049868 (rank : 35) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CLUA1_HUMAN
|
||||||
| NC score | 0.049690 (rank : 36) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96AJ1, O75138, Q65ZA3, Q9H8R4, Q9H8T1 | Gene names | CLUAP1, KIAA0643 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.048980 (rank : 37) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MAP9_HUMAN
|
||||||
| NC score | 0.048749 (rank : 38) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q49MG5, Q4W5I7, Q68DU1, Q9H781, Q9H7B6 | Gene names | MAP9, ASAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
DX26B_HUMAN
|
||||||
| NC score | 0.047436 (rank : 39) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5JSJ4, Q5CZA2, Q6IPS3, Q6ZTU5, Q6ZWE4 | Gene names | DDX26B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
K1383_HUMAN
|
||||||
| NC score | 0.045989 (rank : 40) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2G4, Q58EZ9, Q5VV83 | Gene names | KIAA1383 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1383. | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.045970 (rank : 41) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.045605 (rank : 42) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
TRI44_MOUSE
|
||||||
| NC score | 0.044633 (rank : 43) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QXA7, Q80SV9, Q9JHR8 | Gene names | Trim44, Dipb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB) (Protein Mc7). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.043913 (rank : 44) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
UBP1_HUMAN
|
||||||
| NC score | 0.040021 (rank : 45) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94782, Q9UFR0, Q9UNJ3 | Gene names | USP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 1 (EC 3.1.2.15) (Ubiquitin thioesterase 1) (Ubiquitin-specific-processing protease 1) (Deubiquitinating enzyme 1) (hUBP). | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.037282 (rank : 46) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
NPM_MOUSE
|
||||||
| NC score | 0.036720 (rank : 47) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61937 | Gene names | Npm1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleophosmin (NPM) (Nucleolar phosphoprotein B23) (Numatrin) (Nucleolar protein NO38). | |||||
|
NUCL_HUMAN
|
||||||
| NC score | 0.036642 (rank : 48) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P19338, Q53SK1, Q8NB06 | Gene names | NCL | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
APC_MOUSE
|
||||||
| NC score | 0.036464 (rank : 49) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61315, Q62044 | Gene names | Apc | |||
|
Domain Architecture |

|
|||||
| Description | Adenomatous polyposis coli protein (Protein APC) (mAPC). | |||||
|
CDS2_MOUSE
|
||||||
| NC score | 0.035439 (rank : 50) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
ACS2L_MOUSE
|
||||||
| NC score | 0.034735 (rank : 51) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99NB1, Q8K0M6 | Gene names | Acss1, Acas2, Acas2l | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (AceCS2) (Acyl-CoA synthetase short-chain family member 1). | |||||
|
TOP2B_HUMAN
|
||||||
| NC score | 0.034730 (rank : 52) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02880, Q13600, Q9UMG8, Q9UQP8 | Gene names | TOP2B | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-beta (EC 5.99.1.3) (DNA topoisomerase II, beta isozyme). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.034665 (rank : 53) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
TOP2A_HUMAN
|
||||||
| NC score | 0.034386 (rank : 54) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P11388, Q71UN1, Q9HB24, Q9HB25, Q9HB26, Q9UP44, Q9UQP9 | Gene names | TOP2A, TOP2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.034379 (rank : 55) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.033678 (rank : 56) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.033570 (rank : 57) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.033414 (rank : 58) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
CLUA1_MOUSE
|
||||||
| NC score | 0.033114 (rank : 59) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8R3P7, Q6P3Y8 | Gene names | Cluap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Clusterin-associated protein 1. | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.032041 (rank : 60) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.031192 (rank : 61) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
SNIP1_MOUSE
|
||||||
| NC score | 0.030706 (rank : 62) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BIZ6, Q3V106, Q8BIZ4 | Gene names | Snip1 | |||
|
Domain Architecture |

|
|||||
| Description | Smad nuclear-interacting protein 1. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.030470 (rank : 63) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
TOIP2_HUMAN
|
||||||
| NC score | 0.029729 (rank : 64) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NFQ8 | Gene names | TOR1AIP2, LULL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Torsin-1A-interacting protein 2 (Lumenal domain-like LAP1). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.029269 (rank : 65) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.028647 (rank : 66) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.027891 (rank : 67) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
SDCG1_HUMAN
|
||||||
| NC score | 0.027837 (rank : 68) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60524, Q8WW70, Q9NWG1 | Gene names | SDCCAG1 | |||
|
Domain Architecture |

|
|||||
| Description | Serologically defined colon cancer antigen 1 (Antigen NY-CO-1). | |||||
|
ACS2L_HUMAN
|
||||||
| NC score | 0.027542 (rank : 69) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NUB1, Q5TF42, Q8IV99, Q8N234, Q96JI1, Q96JX6, Q9NU28 | Gene names | ACSS1, ACAS2L, KIAA1846 | |||
|
Domain Architecture |

|
|||||
| Description | Acetyl-coenzyme A synthetase 2-like, mitochondrial precursor (EC 6.2.1.1) (Acetate--CoA ligase 2) (Acetyl-CoA synthetase 2) (Acyl- CoA synthetase short-chain family member 1). | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.027306 (rank : 70) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
DX26B_MOUSE
|
||||||
| NC score | 0.026927 (rank : 71) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BND4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DDX26B. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.026476 (rank : 72) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.026202 (rank : 73) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
LIMA1_HUMAN
|
||||||
| NC score | 0.025030 (rank : 74) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.024973 (rank : 75) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
HSP72_MOUSE
|
||||||
| NC score | 0.024816 (rank : 76) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17156 | Gene names | Hspa2, Hcp70.2, Hsp70-2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock protein 70.2). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.023960 (rank : 77) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.023687 (rank : 78) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
HSP72_HUMAN
|
||||||
| NC score | 0.023073 (rank : 79) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54652, Q15508, Q9UE78 | Gene names | HSPA2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock-related 70 kDa protein 2 (Heat shock 70 kDa protein 2). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.022381 (rank : 80) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.020180 (rank : 81) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
FGD6_HUMAN
|
||||||
| NC score | 0.020139 (rank : 82) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 237 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6ZV73, Q6ZR53, Q7Z2Z7, Q96D44, Q9NUR8, Q9P2I5 | Gene names | FGD6, KIAA1362, ZFYVE24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6 (Zinc finger FYVE domain-containing protein 24). | |||||
|
HNRPC_MOUSE
|
||||||
| NC score | 0.018987 (rank : 83) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z204, Q3TLB5, Q501Q3, Q8C2G5, Q99KE2, Q9CQT3, Q9CY83 | Gene names | Hnrpc, Hnrnpc | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoproteins C1/C2 (hnRNP C1 / hnRNP C2). | |||||
|
TBX2_MOUSE
|
||||||
| NC score | 0.018282 (rank : 84) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60707 | Gene names | Tbx2 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX2 (T-box protein 2). | |||||
|
PRGC2_MOUSE
|
||||||
| NC score | 0.016997 (rank : 85) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VHJ7, Q8C1C0 | Gene names | Ppargc1b, Errl1, Pgc1, Pgc1b, Ppargc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta protein) (PGC-1beta) (ERR ligand 1) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.016252 (rank : 86) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
CK060_HUMAN
|
||||||
| NC score | 0.016211 (rank : 87) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQC8 | Gene names | C11orf60, C11orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0360 protein C11orf60. | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.016052 (rank : 88) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
RBM28_MOUSE
|
||||||
| NC score | 0.015997 (rank : 89) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
CF152_HUMAN
|
||||||
| NC score | 0.015726 (rank : 90) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q86VQ0, Q9BWX7 | Gene names | C6orf152 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf152. | |||||
|
WDFY3_HUMAN
|
||||||
| NC score | 0.014754 (rank : 91) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8IZQ1, Q4W5K5, Q6P0Q5, Q8N1T2, Q96BS7, Q96N85, Q9Y2J7 | Gene names | WDFY3, KIAA0993 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Autophagy-linked FYVE protein) (Alfy). | |||||
|
LRC27_MOUSE
|
||||||
| NC score | 0.014706 (rank : 92) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YS5, Q9D6Z2 | Gene names | Lrrc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 27. | |||||
|
WDFY3_MOUSE
|
||||||
| NC score | 0.013948 (rank : 93) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.013522 (rank : 94) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.013089 (rank : 95) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.012600 (rank : 96) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
DZIP3_HUMAN
|
||||||
| NC score | 0.011583 (rank : 97) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.009857 (rank : 98) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.008514 (rank : 99) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.007867 (rank : 100) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.005273 (rank : 101) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
MASTL_MOUSE
|
||||||
| NC score | 0.002336 (rank : 102) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 97 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C0P0, Q5RJW0, Q6NXX9, Q8BVF3, Q9CZH9, Q9D9V0 | Gene names | Mastl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase-like (EC 2.7.11.1). | |||||