Please be patient as the page loads
|
GORS2_MOUSE
|
||||||
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GORS2_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
GORS2_HUMAN
|
||||||
| θ value | 1.15296e-180 (rank : 2) | NC score | 0.966946 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 1.54831e-84 (rank : 3) | NC score | 0.897445 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
GR65_MOUSE
|
||||||
| θ value | 1.71185e-83 (rank : 4) | NC score | 0.913370 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 5) | NC score | 0.123756 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 6) | NC score | 0.104436 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 7) | NC score | 0.095842 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
SYNG_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.110548 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
CF015_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.122060 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.129441 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.045068 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.040443 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 13) | NC score | 0.082751 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.056846 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
CO002_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.076949 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
F120C_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.071361 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX05 | Gene names | FAM120C, CXorf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C (Tumor antigen BJ-HCC-21). | |||||
|
PRAX_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.081614 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.062305 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
CMTA2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.056232 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.103281 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.073093 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SYNG_MOUSE
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.096764 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
PSMD9_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.172105 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
NFRKB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.084960 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.051396 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PSMD9_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.182196 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.016076 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.040960 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.052015 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.049107 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.053031 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.055565 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.029535 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.060465 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
IF35_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.064967 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.010871 (rank : 110) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
F120C_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.065605 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.049973 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SUHW3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.009749 (rank : 111) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
DAB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.042421 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.025947 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.004203 (rank : 115) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.043538 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DTX2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.022195 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |
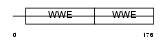
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.024589 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.035165 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.013216 (rank : 104) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.030663 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
ZDH14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.015632 (rank : 100) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQQ1, Q8BNR2, Q8CFN0 | Gene names | Zdhhc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.023081 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.012631 (rank : 107) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
FINC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.016690 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.052812 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
TAU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.044599 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
BAT4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.024566 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |
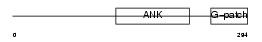
|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.018031 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
HNF1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.030476 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
LAP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.014624 (rank : 102) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
LAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.016229 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.015300 (rank : 101) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.030933 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.066213 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.056376 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
TGIF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.013189 (rank : 105) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0Y1 | Gene names | Tgif2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.012258 (rank : 108) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.048349 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MADCA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.023718 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61826, O35530, Q61278, Q64275, Q8R1M6 | Gene names | Madcam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (mMAdCAM-1). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.013274 (rank : 103) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.025849 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
SC24D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.032778 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
SOX13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.020313 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
TRIO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.005210 (rank : 113) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.049857 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.048422 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.011011 (rank : 109) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.012637 (rank : 106) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.003260 (rank : 116) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.053552 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.022716 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
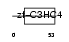
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.037670 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.022005 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
ARHG6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.006297 (rank : 112) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
CASZ1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.023750 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.018055 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MDGA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.005184 (rank : 114) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFP4, Q8NBE3 | Gene names | MDGA1, MAMDC3 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing glycosylphosphatidylinositol anchor protein 1 precursor (MAM domain-containing protein 3) (GPI and MAM protein) (Glycosylphosphatidylinositol-MAM) (GPIM). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.047203 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
SF3A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.052249 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
SF3A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.053426 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.039415 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CIC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.058378 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052931 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.061192 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.056502 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.050980 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.052548 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
LEUK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.053135 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.067477 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050796 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.053740 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.074829 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.064722 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054724 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
PO121_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.055996 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
PRAX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.071749 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050766 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054393 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.070597 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
SF01_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054885 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052394 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051386 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.062315 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.082771 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.070077 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.053114 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.056144 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.067507 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.966946 (rank : 2) | θ value | 1.15296e-180 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GR65_MOUSE
|
||||||
| NC score | 0.913370 (rank : 3) | θ value | 1.71185e-83 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.897445 (rank : 4) | θ value | 1.54831e-84 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
PSMD9_MOUSE
|
||||||
| NC score | 0.182196 (rank : 5) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_HUMAN
|
||||||
| NC score | 0.172105 (rank : 6) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.129441 (rank : 7) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.123756 (rank : 8) | θ value | 1.43324e-05 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CF015_HUMAN
|
||||||
| NC score | 0.122060 (rank : 9) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
SYNG_HUMAN
|
||||||
| NC score | 0.110548 (rank : 10) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UMZ2, Q6ZT17 | Gene names | AP1GBP1, SYNG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.104436 (rank : 11) | θ value | 0.00102713 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.103281 (rank : 12) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SYNG_MOUSE
|
||||||
| NC score | 0.096764 (rank : 13) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5SV85, Q5SV84, Q6PHT6 | Gene names | Ap1gbp1, Syng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP1 subunit gamma-binding protein 1 (Gamma-synergin). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.095842 (rank : 14) | θ value | 0.00134147 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
NFRKB_MOUSE
|
||||||
| NC score | 0.084960 (rank : 15) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6PIJ4, Q8BWV5, Q8K0X6 | Gene names | Nfrkb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.082771 (rank : 16) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.082751 (rank : 17) | θ value | 0.0736092 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
PRAX_MOUSE
|
||||||
| NC score | 0.081614 (rank : 18) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O55103, O55104 | Gene names | Prx | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
CO002_HUMAN
|
||||||
| NC score | 0.076949 (rank : 19) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZP6 | Gene names | C15orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C15orf2. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.074829 (rank : 20) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.073093 (rank : 21) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.071749 (rank : 22) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
F120C_HUMAN
|
||||||
| NC score | 0.071361 (rank : 23) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NX05 | Gene names | FAM120C, CXorf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C (Tumor antigen BJ-HCC-21). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.070597 (rank : 24) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.070077 (rank : 25) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.067507 (rank : 26) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.067477 (rank : 27) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.066213 (rank : 28) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
F120C_MOUSE
|
||||||
| NC score | 0.065605 (rank : 29) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
IF35_MOUSE
|
||||||
| NC score | 0.064967 (rank : 30) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DCH4 | Gene names | Eif3s5 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 5 (eIF-3 epsilon) (eIF3 p47 subunit) (eIF3f). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.064722 (rank : 31) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.062315 (rank : 32) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.062305 (rank : 33) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.061192 (rank : 34) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.060465 (rank : 35) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.058378 (rank : 36) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.056846 (rank : 37) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.056502 (rank : 38) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.056376 (rank : 39) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
CMTA2_HUMAN
|
||||||
| NC score | 0.056232 (rank : 40) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94983, Q7Z6M8, Q8N3V0, Q8NDG4, Q96G17 | Gene names | CAMTA2, KIAA0909 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 2. | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.056144 (rank : 41) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.055996 (rank : 42) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.055565 (rank : 43) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
SF01_HUMAN
|
||||||
| NC score | 0.054885 (rank : 44) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15637, Q14818, Q14819, Q15913, Q8IY00, Q92744, Q92745, Q969H7, Q9BW01, Q9UEI0 | Gene names | SF1, ZFM1, ZNF162 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 1 (Zinc finger protein 162) (Transcription factor ZFM1) (Zinc finger gene in MEN1 locus) (Mammalian branch point-binding protein mBBP) (BBP). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.054724 (rank : 45) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.054393 (rank : 46) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.053740 (rank : 47) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.053552 (rank : 48) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
SF3A1_MOUSE
|
||||||
| NC score | 0.053426 (rank : 49) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8K4Z5, Q8C0M7, Q8C128, Q8C175, Q921T3 | Gene names | Sf3a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 3 subunit 1 (SF3a120). | |||||
|
LEUK_HUMAN
|
||||||
| NC score | 0.053135 (rank : 50) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P16150 | Gene names | SPN, CD43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukosialin precursor (Leukocyte sialoglycoprotein) (Sialophorin) (Galactoglycoprotein) (GALGP) (CD43 antigen). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.053114 (rank : 51) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.053031 (rank : 52) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.052931 (rank : 53) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.052812 (rank : 54) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.052548 (rank : 55) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.052394 (rank : 56) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A1_HUMAN
|
||||||
| NC score | 0.052249 (rank : 57) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q15459 | Gene names | SF3A1, SAP114 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3 subunit 1 (Spliceosome-associated protein 114) (SAP 114) (SF3a120). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.052015 (rank : 58) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.051396 (rank : 59) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.051386 (rank : 60) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.050980 (rank : 61) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.050796 (rank : 62) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.050766 (rank : 63) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.049973 (rank : 64) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.049857 (rank : 65) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.049107 (rank : 66) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.048422 (rank : 67) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.048349 (rank : 68) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.047203 (rank : 69) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.045068 (rank : 70) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.044599 (rank : 71) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.043538 (rank : 72) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DAB1_HUMAN
|
||||||
| NC score | 0.042421 (rank : 73) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75553, Q9NYA8 | Gene names | DAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 1. | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.040960 (rank : 74) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.040443 (rank : 75) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.039415 (rank : 76) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.037670 (rank : 77) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.035165 (rank : 78) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
SC24D_HUMAN
|
||||||
| NC score | 0.032778 (rank : 79) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94855 | Gene names | SEC24D, KIAA0755 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24D (SEC24-related protein D). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.030933 (rank : 80) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.030663 (rank : 81) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
HNF1A_MOUSE
|
||||||
| NC score | 0.030476 (rank : 82) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P22361 | Gene names | Tcf1, Hnf-1, Hnf-1a, Hnf1a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-alpha (HNF-1A) (Liver-specific transcription factor LF-B1) (LFB1). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.029535 (rank : 83) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.025947 (rank : 84) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.025849 (rank : 85) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.024589 (rank : 86) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
BAT4_HUMAN
|
||||||
| NC score | 0.024566 (rank : 87) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95872 | Gene names | BAT4, G5 | |||
|
Domain Architecture |

|
|||||
| Description | Protein BAT4 (HLA-B-associated transcript 4) (G5 protein). | |||||
|
CASZ1_HUMAN
|
||||||
| NC score | 0.023750 (rank : 88) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 473 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86V15, Q5T9S1, Q8WX49, Q8WX50, Q9BT16, Q9NXC6 | Gene names | CASZ1, CST, ZNF693 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Castor homolog 1 zinc finger protein (Castor-related protein) (Zinc finger protein 693). | |||||
|
MADCA_MOUSE
|
||||||
| NC score | 0.023718 (rank : 89) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61826, O35530, Q61278, Q64275, Q8R1M6 | Gene names | Madcam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucosal addressin cell adhesion molecule 1 precursor (MAdCAM-1) (mMAdCAM-1). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.023081 (rank : 90) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.022716 (rank : 91) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
DTX2_MOUSE
|
||||||
| NC score | 0.022195 (rank : 92) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.022005 (rank : 93) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
SOX13_HUMAN
|
||||||
| NC score | 0.020313 (rank : 94) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UN79, O95275, O95826, Q9UHW7 | Gene names | SOX13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein (Type 1 diabetes autoantigen ICA12) (Islet cell antigen 12). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.018055 (rank : 95) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.018031 (rank : 96) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.016690 (rank : 97) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
LAP4_MOUSE
|
||||||
| NC score | 0.016229 (rank : 98) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80U72, Q6P5H7, Q7TPH8, Q80VB1, Q8CI48, Q8VII1, Q922S3 | Gene names | Scrib, Kiaa0147, Lap4, Scrib1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.016076 (rank : 99) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZDH14_MOUSE
|
||||||
| NC score | 0.015632 (rank : 100) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQQ1, Q8BNR2, Q8CFN0 | Gene names | Zdhhc14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.015300 (rank : 101) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
LAP4_HUMAN
|
||||||
| NC score | 0.014624 (rank : 102) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 608 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14160, Q6P496, Q7Z5D1, Q8WWV8, Q96C69, Q96GG1 | Gene names | SCRIB, CRIB1, KIAA0147, LAP4, SCRB1, VARTUL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP4 (Protein scribble homolog) (hScrib). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.013274 (rank : 103) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.013216 (rank : 104) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
TGIF2_MOUSE
|
||||||
| NC score | 0.013189 (rank : 105) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C0Y1 | Gene names | Tgif2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein TGIF2 (TGFB-induced factor 2) (5'-TG-3'-interacting factor 2) (TGF(beta)-induced transcription factor 2). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.012637 (rank : 106) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.012631 (rank : 107) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.012258 (rank : 108) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.011011 (rank : 109) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.010871 (rank : 110) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
SUHW3_HUMAN
|
||||||
| NC score | 0.009749 (rank : 111) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
ARHG6_HUMAN
|
||||||
| NC score | 0.006297 (rank : 112) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15052, Q15396, Q5JQ66, Q86XH0 | Gene names | ARHGEF6, COOL2, KIAA0006, PIXA | |||
|
Domain Architecture |

|
|||||
| Description | Rho guanine nucleotide exchange factor 6 (Rac/Cdc42 guanine nucleotide exchange factor 6) (PAK-interacting exchange factor alpha) (Alpha-Pix) (COOL-2). | |||||
|
TRIO_HUMAN
|
||||||
| NC score | 0.005210 (rank : 113) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1580 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O75962, Q13458 | Gene names | TRIO | |||
|
Domain Architecture |

|
|||||
| Description | Triple functional domain protein (PTPRF-interacting protein). | |||||
|
MDGA1_HUMAN
|
||||||
| NC score | 0.005184 (rank : 114) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFP4, Q8NBE3 | Gene names | MDGA1, MAMDC3 | |||
|
Domain Architecture |

|
|||||
| Description | MAM domain-containing glycosylphosphatidylinositol anchor protein 1 precursor (MAM domain-containing protein 3) (GPI and MAM protein) (Glycosylphosphatidylinositol-MAM) (GPIM). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.004203 (rank : 115) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
GLI3_MOUSE
|
||||||
| NC score | 0.003260 (rank : 116) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||