Please be patient as the page loads
|
CF015_HUMAN
|
||||||
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CF015_HUMAN
|
||||||
| θ value | 2.41524e-162 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 2) | NC score | 0.122060 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 3) | NC score | 0.150289 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
PRIO_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.209499 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04156, O60489, P78446, Q15216, Q15221, Q8TBG0, Q96E70, Q9UP19 | Gene names | PRNP, PRP | |||
|
Domain Architecture |
|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (ASCR) (CD230 antigen). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.068679 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.162403 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.159453 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
K1543_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.103686 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
PRIO_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.205504 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04925 | Gene names | Prnp, Prn-p, Prp | |||
|
Domain Architecture |
|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (CD230 antigen). | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.072701 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.054922 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
ADAM9_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.020175 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61072, Q60618, Q61853, Q80U94 | Gene names | Adam9, Kiaa0021, Mdc9, Mltng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma). | |||||
|
ASPP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.052270 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
TTDN1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.091976 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAP9 | Gene names | TTDN1, C7orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TTD non-photosensitive 1 protein. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.070495 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
FUBP2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.052252 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.079083 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.028547 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.084442 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
ASPP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.051647 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.034922 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CN032_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.077277 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
RPGF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.042000 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.056185 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
CT128_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.054551 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.033553 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
PRR13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.080209 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.072752 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.032581 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.006609 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.060853 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.058437 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RECQ4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.028119 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.023292 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
COFA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.045115 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
SFR17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.047978 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
COAA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.025145 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
COFA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.044157 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.041117 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DAZP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.028752 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DAZP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.029711 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |
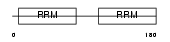
|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.046640 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.100851 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.037203 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.042974 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.028342 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
GGN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.046447 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.049591 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
ACK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.005016 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
CE016_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.026412 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.015642 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.037095 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.031158 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
HNRL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.023877 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
K1683_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.032744 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
KSR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.004562 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.016005 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.018462 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.036055 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.018384 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
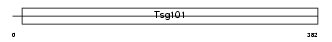
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.052743 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GORS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.067016 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GR65_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.051037 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
K1543_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.068082 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MISS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.059270 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054531 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.053466 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051938 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
CF015_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.41524e-162 (rank : 1) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q6UXA7, Q5SQ81, Q86Z05, Q9UIG3 | Gene names | C6orf15, STG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf15 precursor (Protein STG). | |||||
|
PRIO_HUMAN
|
||||||
| NC score | 0.209499 (rank : 2) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P04156, O60489, P78446, Q15216, Q15221, Q8TBG0, Q96E70, Q9UP19 | Gene names | PRNP, PRP | |||
|
Domain Architecture |

|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (ASCR) (CD230 antigen). | |||||
|
PRIO_MOUSE
|
||||||
| NC score | 0.205504 (rank : 3) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04925 | Gene names | Prnp, Prn-p, Prp | |||
|
Domain Architecture |

|
|||||
| Description | Major prion protein precursor (PrP) (PrP27-30) (PrP33-35C) (CD230 antigen). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.162403 (rank : 4) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.159453 (rank : 5) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.150289 (rank : 6) | θ value | 0.0148317 (rank : 3) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.122060 (rank : 7) | θ value | 0.00665767 (rank : 2) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.103686 (rank : 8) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.100851 (rank : 9) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
TTDN1_HUMAN
|
||||||
| NC score | 0.091976 (rank : 10) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAP9 | Gene names | TTDN1, C7orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TTD non-photosensitive 1 protein. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.084442 (rank : 11) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.080209 (rank : 12) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.079083 (rank : 13) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.077277 (rank : 14) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.072752 (rank : 15) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.072701 (rank : 16) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.070495 (rank : 17) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.068679 (rank : 18) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.068082 (rank : 19) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.067016 (rank : 20) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.060853 (rank : 21) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.059270 (rank : 22) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.058437 (rank : 23) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.056185 (rank : 24) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.054922 (rank : 25) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CT128_HUMAN
|
||||||
| NC score | 0.054551 (rank : 26) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.054531 (rank : 27) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.053466 (rank : 28) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.052743 (rank : 29) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ASPP1_HUMAN
|
||||||
| NC score | 0.052270 (rank : 30) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 956 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96KQ4, O94870 | Gene names | PPP1R13B, ASPP1, KIAA0771 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
FUBP2_HUMAN
|
||||||
| NC score | 0.052252 (rank : 31) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92945, O00301, Q9UNT5, Q9UQH5 | Gene names | KHSRP, FUBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type-splicing regulatory protein) (KSRP) (p75). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.051938 (rank : 32) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ASPP1_MOUSE
|
||||||
| NC score | 0.051647 (rank : 33) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62415 | Gene names | Ppp1r13b, Aspp1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-stimulating of p53 protein 1 (Protein phosphatase 1 regulatory subunit 13B). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.051037 (rank : 34) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.049591 (rank : 35) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.047978 (rank : 36) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.046640 (rank : 37) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.046447 (rank : 38) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
COFA1_HUMAN
|
||||||
| NC score | 0.045115 (rank : 39) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P39059, Q5T6J4, Q9Y4W4 | Gene names | COL15A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV) (Restin) (Related to endostatin)]. | |||||
|
COFA1_MOUSE
|
||||||
| NC score | 0.044157 (rank : 40) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35206, Q9EQD9 | Gene names | Col15a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XV) chain precursor [Contains: Endostatin (Endostatin-XV)]. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.042974 (rank : 41) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
RPGF1_HUMAN
|
||||||
| NC score | 0.042000 (rank : 42) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13905 | Gene names | RAPGEF1, GRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 1 (Guanine nucleotide-releasing factor 2) (C3G protein) (CRK SH3-binding GNRP). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.041117 (rank : 43) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.037203 (rank : 44) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.037095 (rank : 45) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.036055 (rank : 46) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.034922 (rank : 47) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.033553 (rank : 48) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.032744 (rank : 49) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.032581 (rank : 50) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.031158 (rank : 51) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
DAZP1_MOUSE
|
||||||
| NC score | 0.029711 (rank : 52) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JII5 | Gene names | Dazap1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
DAZP1_HUMAN
|
||||||
| NC score | 0.028752 (rank : 53) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EP5, Q96MJ3, Q9NRR9 | Gene names | DAZAP1 | |||
|
Domain Architecture |

|
|||||
| Description | DAZ-associated protein 1 (Deleted in azoospermia-associated protein 1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.028547 (rank : 54) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.028342 (rank : 55) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
RECQ4_HUMAN
|
||||||
| NC score | 0.028119 (rank : 56) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O94761, Q3Y424, Q96DW2, Q96F55 | Gene names | RECQL4, RECQ4 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4) (RecQ4) (RTS). | |||||
|
CE016_HUMAN
|
||||||
| NC score | 0.026412 (rank : 57) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NC24, Q6P4E7, Q6UXY2 | Gene names | C5orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C5orf16. | |||||
|
COAA1_HUMAN
|
||||||
| NC score | 0.025145 (rank : 58) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q03692 | Gene names | COL10A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
HNRL1_HUMAN
|
||||||
| NC score | 0.023877 (rank : 59) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BUJ2, O76022, Q6ZSZ0, Q7L8P4, Q8N6Z4, Q96G37, Q9HAL3, Q9UG75 | Gene names | HNRPUL1, E1BAP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U-like protein 1 (Adenovirus early region 1B-associated protein 5) (E1B-55 kDa-associated protein 5) (E1B-AP5). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.023292 (rank : 60) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ADAM9_MOUSE
|
||||||
| NC score | 0.020175 (rank : 61) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61072, Q60618, Q61853, Q80U94 | Gene names | Adam9, Kiaa0021, Mdc9, Mltng | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAM 9 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 9) (Metalloprotease/disintegrin/cysteine-rich protein 9) (Myeloma cell metalloproteinase) (Meltrin gamma). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.018462 (rank : 62) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.018384 (rank : 63) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.016005 (rank : 64) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.015642 (rank : 65) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.006609 (rank : 66) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
ACK1_HUMAN
|
||||||
| NC score | 0.005016 (rank : 67) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 1140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q07912, Q8N6U7, Q96H59 | Gene names | TNK2, ACK1 | |||
|
Domain Architecture |

|
|||||
| Description | Activated CDC42 kinase 1 (EC 2.7.10.2) (ACK-1) (Tyrosine kinase non- receptor protein 2). | |||||
|
KSR1_HUMAN
|
||||||
| NC score | 0.004562 (rank : 68) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 60 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IVT5, Q13476 | Gene names | KSR1, KSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinase suppressor of ras-1 (Kinase suppressor of ras). | |||||