Please be patient as the page loads
|
PSMD9_HUMAN
|
||||||
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PSMD9_HUMAN
|
||||||
| θ value | 1.99884e-124 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_MOUSE
|
||||||
| θ value | 1.75921e-104 (rank : 2) | NC score | 0.986541 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 3) | NC score | 0.214607 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
NHRF2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 4) | NC score | 0.173624 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
GR65_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.209755 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
INADL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 6) | NC score | 0.096926 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
NHERF_HUMAN
|
||||||
| θ value | 0.125558 (rank : 7) | NC score | 0.150138 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |
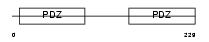
|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
NHRF2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 8) | NC score | 0.158925 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
PDZD1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.129659 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
USH1C_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.123569 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
WHRN_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.118949 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
GORS2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.173901 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.172105 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.076735 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.026366 (rank : 54) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
CCHCR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.031713 (rank : 53) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.109692 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
PDZD1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.129694 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.021991 (rank : 55) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
PDLI1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.061247 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |
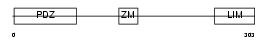
|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
INADL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.096746 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
HTRA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.054408 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.011626 (rank : 60) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.016000 (rank : 57) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
WHRN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.101936 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.015194 (rank : 58) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
RPGF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.069480 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
RPGF6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.069679 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.040416 (rank : 51) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
PTN13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.036250 (rank : 52) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
AR13B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.008012 (rank : 61) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.012255 (rank : 59) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FRAT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.016432 (rank : 56) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |
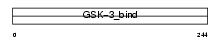
|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
HTRA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.042417 (rank : 50) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
TX1B3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.085152 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.085124 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
GRASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.050918 (rank : 48) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
GRASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.050840 (rank : 49) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
GRIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.056430 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
GRIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.057117 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
LIN7A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.054497 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.054469 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
LIN7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.054034 (rank : 39) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
LIN7B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.053718 (rank : 43) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
LIN7C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.053255 (rank : 45) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.053255 (rank : 46) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.053835 (rank : 41) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.054069 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.074494 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.071225 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
NHERF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.117303 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.054988 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.053726 (rank : 42) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
PARD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.053978 (rank : 40) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
PDZ11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.056519 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZ11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.057518 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
PDZD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.098002 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
PDZD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.102464 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
PDZD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.084382 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
PSCBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.052588 (rank : 47) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
PSCBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.053666 (rank : 44) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VY6, Q3TZH1, Q8R4T4, Q9QZA9 | Gene names | Pscdbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (Cbp). | |||||
|
PSMD9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.99884e-124 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00233, Q9BQ42 | Gene names | PSMD9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
PSMD9_MOUSE
|
||||||
| NC score | 0.986541 (rank : 2) | θ value | 1.75921e-104 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9CR00, Q3TG66 | Gene names | Psmd9 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 9 (26S proteasome regulatory subunit p27). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.214607 (rank : 3) | θ value | 0.0113563 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
GR65_MOUSE
|
||||||
| NC score | 0.209755 (rank : 4) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.173901 (rank : 5) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
NHRF2_MOUSE
|
||||||
| NC score | 0.173624 (rank : 6) | θ value | 0.0193708 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JHL1, Q3TDR3, Q8BGL9, Q8BW05, Q9DCR6 | Gene names | Slc9a3r2, Nherf2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2) (Octs2). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.172105 (rank : 7) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
NHRF2_HUMAN
|
||||||
| NC score | 0.158925 (rank : 8) | θ value | 0.125558 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q15599, O00272, O00556 | Gene names | SLC9A3R2, NHERF2 | |||
|
Domain Architecture |

|
|||||
| Description | Na(+)/H(+) exchange regulatory cofactor NHE-RF2 (NHERF-2) (Tyrosine kinase activator protein 1) (TKA-1) (SRY-interacting protein 1) (SIP- 1) (Solute carrier family 9 isoform A3 regulatory factor 2) (NHE3 kinase A regulatory protein E3KARP) (Sodium-hydrogen exchanger regulatory factor 2). | |||||
|
NHERF_HUMAN
|
||||||
| NC score | 0.150138 (rank : 9) | θ value | 0.125558 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14745, O43552, Q86WQ5 | Gene names | SLC9A3R1, NHERF | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
PDZD1_HUMAN
|
||||||
| NC score | 0.129694 (rank : 10) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5T2W1, O60450, Q5T5P6, Q9BQ41 | Gene names | PDZK1, CAP70, NHERF3, PDZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
PDZD1_MOUSE
|
||||||
| NC score | 0.129659 (rank : 11) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIL4, Q8CDP5, Q8R4G2, Q9CQ72 | Gene names | Pdzk1, Cap70, Nherf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 1 (CFTR-associated protein of 70 kDa) (Na/Pi cotransporter C-terminal-associated protein) (NaPi-Cap1) (Na(+)/H(+) exchanger regulatory factor 3) (Sodium-hydrogen exchanger regulatory factor 3). | |||||
|
USH1C_HUMAN
|
||||||
| NC score | 0.123569 (rank : 12) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y6N9, Q96B29, Q9UM04, Q9UM17, Q9UPC3 | Gene names | USH1C, AIE75 | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein) (Autoimmune enteropathy- related antigen AIE-75) (Antigen NY-CO-38/NY-CO-37) (PDZ-73 protein) (NY-REN-3 antigen). | |||||
|
WHRN_MOUSE
|
||||||
| NC score | 0.118949 (rank : 13) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80VW5, Q80TC2, Q80VW4 | Gene names | Whrn, Kiaa1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin. | |||||
|
NHERF_MOUSE
|
||||||
| NC score | 0.117303 (rank : 14) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P70441 | Gene names | Slc9a3r1, Nherf | |||
|
Domain Architecture |

|
|||||
| Description | Ezrin-radixin-moesin-binding phosphoprotein 50 (EBP50) (Na(+)/H(+) exchange regulatory cofactor NHE-RF) (NHERF-1) (Regulatory cofactor of Na(+)/H(+) exchanger) (Sodium-hydrogen exchanger regulatory factor 1) (Solute carrier family 9 isoform 3 regulatory factor 1). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.109692 (rank : 15) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
PDZD3_MOUSE
|
||||||
| NC score | 0.102464 (rank : 16) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q99MJ6, Q8BWE5 | Gene names | Pdzd3, Pdzk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Natrium-phosphate cotransporter IIa C-terminal-associated protein 2) (NaPi-Cap2). | |||||
|
WHRN_HUMAN
|
||||||
| NC score | 0.101936 (rank : 17) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9P202, Q96MZ9, Q9H9F4, Q9UFZ3 | Gene names | WHRN, DFNB31, KIAA1526 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Whirlin (Autosomal recessive deafness type 31 protein). | |||||
|
PDZD3_HUMAN
|
||||||
| NC score | 0.098002 (rank : 18) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UT5, Q8N6R4, Q8NAW7, Q8NEX7, Q9H5Z3 | Gene names | PDZD3, IKEPP, PDZK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 3 (PDZ domain-containing protein 2) (Intestinal and kidney-enriched PDZ protein) (Protein DLNB27). | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.096926 (rank : 19) | θ value | 0.125558 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
INADL_MOUSE
|
||||||
| NC score | 0.096746 (rank : 20) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q63ZW7, O70471, Q5PRG3, Q6P6J1, Q80YR8, Q8BPB9 | Gene names | Inadl, Cipp, Patj | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (Pals1-associated tight junction protein) (Protein associated to tight junctions) (Channel-interacting PDZ domain-containing protein). | |||||
|
TX1B3_HUMAN
|
||||||
| NC score | 0.085152 (rank : 21) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O14907, Q7LCQ4 | Gene names | TAX1BP3, TIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1) (Glutaminase-interacting protein 3). | |||||
|
TX1B3_MOUSE
|
||||||
| NC score | 0.085124 (rank : 22) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBG9 | Gene names | Tax1bp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 3 (Tax interaction protein 1) (TIP-1). | |||||
|
PDZD7_HUMAN
|
||||||
| NC score | 0.084382 (rank : 23) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H5P4, Q8N321 | Gene names | PDZD7, PDZK7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 7. | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.076735 (rank : 24) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.074494 (rank : 25) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
MPDZ_MOUSE
|
||||||
| NC score | 0.071225 (rank : 26) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VBX6, O08783, Q6P7U4, Q80ZY8, Q8BKJ1, Q8C0H8, Q8VBV5, Q8VBY0, Q9Z1K3 | Gene names | Mpdz, Mupp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ domain protein 1). | |||||
|
RPGF6_HUMAN
|
||||||
| NC score | 0.069679 (rank : 27) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8TEU7, Q8NI21, Q8TEU6, Q96PC1 | Gene names | RAPGEF6, PDZGEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Rap guanine nucleotide exchange factor 6 (PDZ domain-containing guanine nucleotide exchange factor 2) (PDZ-GEF2) (RA-GEF-2). | |||||
|
RPGF2_HUMAN
|
||||||
| NC score | 0.069480 (rank : 28) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y4G8 | Gene names | RAPGEF2, KIAA0313, NRAPGEP, PDZGEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rap guanine nucleotide exchange factor 2 (Neural RAP guanine nucleotide exchange protein) (nRap GEP) (PDZ domain-containing guanine nucleotide exchange factor 1) (PDZ-GEF1) (RA-GEF). | |||||
|
PDLI1_HUMAN
|
||||||
| NC score | 0.061247 (rank : 29) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
PDZ11_MOUSE
|
||||||
| NC score | 0.057518 (rank : 30) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9CZG9 | Gene names | Pdzd11, Pdzk11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
GRIP1_MOUSE
|
||||||
| NC score | 0.057117 (rank : 31) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q925T6, Q8BLQ3, Q8C0T3, Q925T5, Q925T7 | Gene names | Grip1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
PDZ11_HUMAN
|
||||||
| NC score | 0.056519 (rank : 32) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5EBL8, Q6UWE1, Q9P0Q1 | Gene names | PDZD11, PDZK11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 11. | |||||
|
GRIP1_HUMAN
|
||||||
| NC score | 0.056430 (rank : 33) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.054988 (rank : 34) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
LIN7A_HUMAN
|
||||||
| NC score | 0.054497 (rank : 35) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14910, Q6LES3, Q7LDS4 | Gene names | LIN7A, MALS1, VELI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (hLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein) (Tax interaction protein 33) (TIP-33). | |||||
|
LIN7A_MOUSE
|
||||||
| NC score | 0.054469 (rank : 36) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8JZS0 | Gene names | Lin7a, Mals1, Veli1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog A (Lin-7A) (mLin-7) (Mammalian lin-seven protein 1) (MALS-1) (Vertebrate lin-7 homolog 1) (Veli-1 protein). | |||||
|
HTRA1_HUMAN
|
||||||
| NC score | 0.054408 (rank : 37) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92743, Q9UNS5 | Gene names | HTRA1, HTRA, PRSS11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-) (L56). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.054069 (rank : 38) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
LIN7B_HUMAN
|
||||||
| NC score | 0.054034 (rank : 39) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9HAP6 | Gene names | LIN7B, MALS2, VELI2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (hLin7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein) (hVeli2). | |||||
|
PARD3_MOUSE
|
||||||
| NC score | 0.053978 (rank : 40) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99NH2 | Gene names | Pard3, Par3 | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (Ephrin-interacting protein) (PHIP). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.053835 (rank : 41) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.053726 (rank : 42) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
LIN7B_MOUSE
|
||||||
| NC score | 0.053718 (rank : 43) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88951 | Gene names | Lin7b, Mals2, Veli2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog B (Lin-7B) (Mammalian lin-seven protein 2) (MALS-2) (Vertebrate lin-7 homolog 2) (Veli-2 protein). | |||||
|
PSCBP_MOUSE
|
||||||
| NC score | 0.053666 (rank : 44) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91VY6, Q3TZH1, Q8R4T4, Q9QZA9 | Gene names | Pscdbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (Cbp). | |||||
|
LIN7C_HUMAN
|
||||||
| NC score | 0.053255 (rank : 45) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NUP9 | Gene names | LIN7C, MALS3, VELI3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
LIN7C_MOUSE
|
||||||
| NC score | 0.053255 (rank : 46) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88952 | Gene names | Lin7c, Mals3, Veli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lin-7 homolog C (Lin-7C) (mLin7C) (Mammalian lin-seven protein 3) (MALS-3) (Vertebrate lin-7 homolog 3) (Veli-3 protein). | |||||
|
PSCBP_HUMAN
|
||||||
| NC score | 0.052588 (rank : 47) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O60759, Q15630, Q8NE32 | Gene names | PSCDBP, CYTIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology Sec7 and coiled-coil domains-binding protein (Cytohesin-binding protein HE) (CYBR) (Cytohesin binder and regulator) (Cytohesin-interacting protein). | |||||
|
GRASP_HUMAN
|
||||||
| NC score | 0.050918 (rank : 48) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7Z6J2, Q6PIF8, Q7Z741 | Gene names | GRASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
GRASP_MOUSE
|
||||||
| NC score | 0.050840 (rank : 49) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
HTRA1_MOUSE
|
||||||
| NC score | 0.042417 (rank : 50) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R118 | Gene names | Htra1, Htra, Prss11 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease HTRA1 precursor (EC 3.4.21.-). | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.040416 (rank : 51) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
PTN13_HUMAN
|
||||||
| NC score | 0.036250 (rank : 52) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q12923, Q15159, Q15263, Q15264, Q15265, Q15674, Q16826, Q8IWH7, Q9NYN9 | Gene names | PTPN13, PNP1, PTP1E, PTPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1E) (PTP-E1) (hPTPE1) (PTP-BAS) (Protein-tyrosine phosphatase PTPL1) (Fas-associated protein-tyrosine phosphatase 1) (FAP-1). | |||||
|
CCHCR_HUMAN
|
||||||
| NC score | 0.031713 (rank : 53) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 717 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TD31, Q9NRK8, Q9NWY9, Q9NXJ4, Q9NXK3, Q9Y6W1, Q9Y6W2 | Gene names | CCHCR1, C6orf18, HCR | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein) (Putative gene 8 protein) (Pg8). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.026366 (rank : 54) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.021991 (rank : 55) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
FRAT1_MOUSE
|
||||||
| NC score | 0.016432 (rank : 56) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P70339 | Gene names | Frat1 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene FRAT1 (Frequently rearranged in advanced T-cell lymphomas) (Frat-1). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.016000 (rank : 57) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.015194 (rank : 58) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.012255 (rank : 59) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.011626 (rank : 60) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
AR13B_MOUSE
|
||||||
| NC score | 0.008012 (rank : 61) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q640N2 | Gene names | Arl13b, Arl2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-like protein 13B (ADP-ribosylation factor-like protein 2-like 1) (ARL2-like protein 1). | |||||