Please be patient as the page loads
|
ENPP2_MOUSE
|
||||||
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ENPP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.975756 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.974011 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.997442 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.977117 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_MOUSE
|
||||||
| θ value | 8.02596e-49 (rank : 6) | NC score | 0.867167 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_HUMAN
|
||||||
| θ value | 1.788e-48 (rank : 7) | NC score | 0.867380 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 9.81109e-47 (rank : 8) | NC score | 0.875613 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP6_MOUSE
|
||||||
| θ value | 3.15612e-45 (rank : 9) | NC score | 0.852084 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_HUMAN
|
||||||
| θ value | 5.38352e-45 (rank : 10) | NC score | 0.852961 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 11) | NC score | 0.202400 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 12) | NC score | 0.179723 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 13) | NC score | 0.158851 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 0.00102713 (rank : 14) | NC score | 0.165536 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
PP11_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 15) | NC score | 0.300217 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 16) | NC score | 0.051775 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
PIGO_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.165290 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 18) | NC score | 0.159368 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.080379 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
TENA_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 20) | NC score | 0.048490 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.036301 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ARSA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.051361 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15289, Q6ICI5, Q96CJ0 | Gene names | ARSA | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase) [Contains: Arylsulfatase A component B; Arylsulfatase A component C]. | |||||
|
ADA28_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.026614 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKQ2, Q9Y339, Q9Y3S0 | Gene names | ADAM28, MDCL | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein-L) (MDC-L) (eMDC II) (ADAM23). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.047185 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
TENA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 25) | NC score | 0.047347 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TECTA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.033545 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.043353 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FSTL5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.029745 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 29) | NC score | 0.099604 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.040280 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 31) | NC score | 0.040433 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
TENR_HUMAN
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.045526 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 0.365318 (rank : 33) | NC score | 0.043960 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
TINAG_HUMAN
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.083087 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
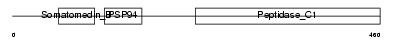
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
ARSA_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.040396 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50428 | Gene names | Arsa, As2 | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase). | |||||
|
CNBP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.041400 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.026130 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.049337 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.058485 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.037553 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.083021 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
FBN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.040391 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
TENX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.039650 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.024949 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
TENN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.035323 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
ARSF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.029261 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54793, Q8TCC5 | Gene names | ARSF | |||
|
Domain Architecture |
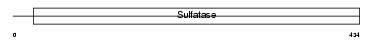
|
|||||
| Description | Arylsulfatase F precursor (EC 3.1.6.-) (ASF). | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.036128 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.032652 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.032097 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
WFDC8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.029104 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |
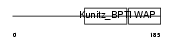
|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
STS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.028335 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50427 | Gene names | Sts | |||
|
Domain Architecture |
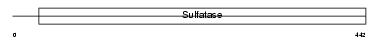
|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
ADAM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.016205 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99965, P78326, Q9UQQ8 | Gene names | ADAM2, FTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 2 precursor (A disintegrin and metalloproteinase domain 2) (Fertilin subunit beta) (PH-30) (PH30). | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.005679 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.022544 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
GRN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.043734 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
KRA57_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.046191 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.013107 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TINAL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.016982 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
ADA28_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.016378 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JLN6, Q8K5D2, Q8K5D3 | Gene names | Adam28 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Thymic epithelial cell-ADAM) (TECADAM). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.028858 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
AS3MT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.009580 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBK9 | Gene names | AS3MT, CYT19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arsenite methyltransferase (EC 2.1.1.137) (S-adenosyl-L- methionine:arsenic(III) methyltransferase) (Methylarsonite methyltransferase). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.039359 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.035784 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.030671 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.031470 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SEZ6L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.006426 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.055659 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
PIGG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.131506 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
PIGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052963 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
PIGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.061669 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.997442 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.977117 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.975756 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.974011 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.875613 (rank : 6) | θ value | 9.81109e-47 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP5_HUMAN
|
||||||
| NC score | 0.867380 (rank : 7) | θ value | 1.788e-48 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| NC score | 0.867167 (rank : 8) | θ value | 8.02596e-49 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP6_HUMAN
|
||||||
| NC score | 0.852961 (rank : 9) | θ value | 5.38352e-45 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| NC score | 0.852084 (rank : 10) | θ value | 3.15612e-45 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
PP11_HUMAN
|
||||||
| NC score | 0.300217 (rank : 11) | θ value | 0.00298849 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.202400 (rank : 12) | θ value | 3.08544e-08 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.179723 (rank : 13) | θ value | 7.59969e-07 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.165536 (rank : 14) | θ value | 0.00102713 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
PIGO_HUMAN
|
||||||
| NC score | 0.165290 (rank : 15) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| NC score | 0.159368 (rank : 16) | θ value | 0.0113563 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.158851 (rank : 17) | θ value | 0.00102713 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
PIGG_HUMAN
|
||||||
| NC score | 0.131506 (rank : 18) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.099604 (rank : 19) | θ value | 0.279714 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.083087 (rank : 20) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.083021 (rank : 21) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.080379 (rank : 22) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.061669 (rank : 23) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.058485 (rank : 24) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.055659 (rank : 25) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.052963 (rank : 26) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.051775 (rank : 27) | θ value | 0.00509761 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
ARSA_HUMAN
|
||||||
| NC score | 0.051361 (rank : 28) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15289, Q6ICI5, Q96CJ0 | Gene names | ARSA | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase) [Contains: Arylsulfatase A component B; Arylsulfatase A component C]. | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.049337 (rank : 29) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.048490 (rank : 30) | θ value | 0.0193708 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.047347 (rank : 31) | θ value | 0.163984 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.047185 (rank : 32) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.046191 (rank : 33) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.045526 (rank : 34) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.043960 (rank : 35) | θ value | 0.365318 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.043734 (rank : 36) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.043353 (rank : 37) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
CNBP_MOUSE
|
||||||
| NC score | 0.041400 (rank : 38) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53996, Q80Y06, Q8BP23 | Gene names | Cnbp, Cnbp1, Znf9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cellular nucleic acid-binding protein (CNBP) (Zinc finger protein 9). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.040433 (rank : 39) | θ value | 0.365318 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
ARSA_MOUSE
|
||||||
| NC score | 0.040396 (rank : 40) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50428 | Gene names | Arsa, As2 | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase A precursor (EC 3.1.6.8) (ASA) (Cerebroside-sulfatase). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.040391 (rank : 41) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.040280 (rank : 42) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.039650 (rank : 43) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.039359 (rank : 44) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.037553 (rank : 45) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.036301 (rank : 46) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.036128 (rank : 47) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.035784 (rank : 48) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
TENN_HUMAN
|
||||||
| NC score | 0.035323 (rank : 49) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQP3, Q5R360 | Gene names | TNN | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-N precursor (TN-N). | |||||
|
TECTA_MOUSE
|
||||||
| NC score | 0.033545 (rank : 50) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O08523 | Gene names | Tecta | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-tectorin precursor. | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.032652 (rank : 51) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.032097 (rank : 52) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.031470 (rank : 53) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.030671 (rank : 54) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
FSTL5_HUMAN
|
||||||
| NC score | 0.029745 (rank : 55) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N475, Q9NSW7, Q9ULF7 | Gene names | FSTL5, KIAA1263 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 5 precursor (Follistatin-like 5). | |||||
|
ARSF_HUMAN
|
||||||
| NC score | 0.029261 (rank : 56) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P54793, Q8TCC5 | Gene names | ARSF | |||
|
Domain Architecture |

|
|||||
| Description | Arylsulfatase F precursor (EC 3.1.6.-) (ASF). | |||||
|
WFDC8_HUMAN
|
||||||
| NC score | 0.029104 (rank : 57) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IUA0, Q5TDV2, Q96A34 | Gene names | WFDC8, C20orf170, WAP8 | |||
|
Domain Architecture |

|
|||||
| Description | WAP four-disulfide core domain protein 8 precursor (Putative protease inhibitor WAP8). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.028858 (rank : 58) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
STS_MOUSE
|
||||||
| NC score | 0.028335 (rank : 59) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P50427 | Gene names | Sts | |||
|
Domain Architecture |

|
|||||
| Description | Steryl-sulfatase precursor (EC 3.1.6.2) (Steroid sulfatase) (Steryl- sulfate sulfohydrolase) (Arylsulfatase C) (ASC). | |||||
|
ADA28_HUMAN
|
||||||
| NC score | 0.026614 (rank : 60) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKQ2, Q9Y339, Q9Y3S0 | Gene names | ADAM28, MDCL | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein-L) (MDC-L) (eMDC II) (ADAM23). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.026130 (rank : 61) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.024949 (rank : 62) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.022544 (rank : 63) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
TINAL_MOUSE
|
||||||
| NC score | 0.016982 (rank : 64) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99JR5, Q9EQT9 | Gene names | Tinagl1, Arg1, Lcn7, Tinagl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulointerstitial nephritis antigen-like precursor (Androgen- regulated gene 1 protein) (Adrenocortical zonation factor 1) (AZ-1) (Tubulointerstitial nephritis antigen-related protein) (TARP). | |||||
|
ADA28_MOUSE
|
||||||
| NC score | 0.016378 (rank : 65) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JLN6, Q8K5D2, Q8K5D3 | Gene names | Adam28 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 28 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 28) (Thymic epithelial cell-ADAM) (TECADAM). | |||||
|
ADAM2_HUMAN
|
||||||
| NC score | 0.016205 (rank : 66) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q99965, P78326, Q9UQQ8 | Gene names | ADAM2, FTNB | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 2 precursor (A disintegrin and metalloproteinase domain 2) (Fertilin subunit beta) (PH-30) (PH30). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.013107 (rank : 67) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
AS3MT_HUMAN
|
||||||
| NC score | 0.009580 (rank : 68) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBK9 | Gene names | AS3MT, CYT19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arsenite methyltransferase (EC 2.1.1.137) (S-adenosyl-L- methionine:arsenic(III) methyltransferase) (Methylarsonite methyltransferase). | |||||
|
SEZ6L_HUMAN
|
||||||
| NC score | 0.006426 (rank : 69) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BYH1, O95917, Q5THY5, Q6IBZ4, Q6UXD4, Q9NUI3, Q9NUI4, Q9NUI5, Q9Y2E1, Q9Y3J6 | Gene names | SEZ6L, KIAA0927 | |||
|
Domain Architecture |

|
|||||
| Description | Seizure 6-like protein precursor. | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.005679 (rank : 70) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||