Please be patient as the page loads
|
ENPP3_HUMAN
|
||||||
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ENPP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989046 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.988548 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.978302 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.977117 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_MOUSE
|
||||||
| θ value | 1.04338e-64 (rank : 6) | NC score | 0.897369 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 7) | NC score | 0.897903 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP6_HUMAN
|
||||||
| θ value | 3.04986e-48 (rank : 8) | NC score | 0.875867 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| θ value | 3.04986e-48 (rank : 9) | NC score | 0.873390 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 3.37202e-47 (rank : 10) | NC score | 0.891783 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 11) | NC score | 0.190358 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 12) | NC score | 0.169751 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TINAG_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 13) | NC score | 0.094474 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
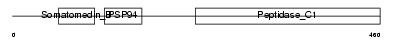
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
PP11_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.291222 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.088094 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
PIGG_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.138166 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.134652 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.101857 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.011498 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.013148 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.029770 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.010289 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.125055 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
ENDD1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.016027 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C522, Q4VA96, Q8C5M1, Q8VE35 | Gene names | Endod1, Kiaa0830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease domain-containing 1 protein precursor (EC 3.1.30.-). | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.009201 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
NELL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.007831 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.007086 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
ITA9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.004050 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
PIGN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.062484 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
SLIT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.002610 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TR4, Q9WVB5 | Gene names | Slit1, Kiaa0813 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1). | |||||
|
ITB6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.005210 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.005935 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PIGN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.070700 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
ZN639_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | -0.001353 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UID6 | Gene names | ZNF639, ZASC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 639 (Zinc finger protein ZASC1) (Zinc finger protein ANC_2H01). | |||||
|
PIGO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.123205 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.116866 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.989046 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.988548 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.978302 (rank : 4) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.977117 (rank : 5) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_HUMAN
|
||||||
| NC score | 0.897903 (rank : 6) | θ value | 1.96772e-63 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_MOUSE
|
||||||
| NC score | 0.897369 (rank : 7) | θ value | 1.04338e-64 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.891783 (rank : 8) | θ value | 3.37202e-47 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP6_HUMAN
|
||||||
| NC score | 0.875867 (rank : 9) | θ value | 3.04986e-48 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| NC score | 0.873390 (rank : 10) | θ value | 3.04986e-48 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
PP11_HUMAN
|
||||||
| NC score | 0.291222 (rank : 11) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.190358 (rank : 12) | θ value | 1.53129e-07 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.169751 (rank : 13) | θ value | 1.29631e-06 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PIGG_HUMAN
|
||||||
| NC score | 0.138166 (rank : 14) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.134652 (rank : 15) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.125055 (rank : 16) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
PIGO_HUMAN
|
||||||
| NC score | 0.123205 (rank : 17) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| NC score | 0.116866 (rank : 18) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.101857 (rank : 19) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.094474 (rank : 20) | θ value | 0.00134147 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.088094 (rank : 21) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.070700 (rank : 22) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.062484 (rank : 23) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.029770 (rank : 24) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
ENDD1_MOUSE
|
||||||
| NC score | 0.016027 (rank : 25) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C522, Q4VA96, Q8C5M1, Q8VE35 | Gene names | Endod1, Kiaa0830 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endonuclease domain-containing 1 protein precursor (EC 3.1.30.-). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.013148 (rank : 26) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.011498 (rank : 27) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.010289 (rank : 28) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.009201 (rank : 29) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.007831 (rank : 30) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.007086 (rank : 31) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.005935 (rank : 32) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.005210 (rank : 33) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITA9_HUMAN
|
||||||
| NC score | 0.004050 (rank : 34) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13797, Q14638 | Gene names | ITGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-9 precursor (Integrin alpha-RLC). | |||||
|
SLIT1_MOUSE
|
||||||
| NC score | 0.002610 (rank : 35) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80TR4, Q9WVB5 | Gene names | Slit1, Kiaa0813 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1). | |||||
|
ZN639_HUMAN
|
||||||
| NC score | -0.001353 (rank : 36) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UID6 | Gene names | ZNF639, ZASC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 639 (Zinc finger protein ZASC1) (Zinc finger protein ANC_2H01). | |||||