Please be patient as the page loads
|
ENPP1_HUMAN
|
||||||
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ENPP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993516 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.975312 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.975756 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.989046 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_MOUSE
|
||||||
| θ value | 1.6097e-65 (rank : 6) | NC score | 0.891055 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 7) | NC score | 0.890968 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP6_HUMAN
|
||||||
| θ value | 6.56568e-51 (rank : 8) | NC score | 0.868958 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| θ value | 3.04986e-48 (rank : 9) | NC score | 0.866532 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP7_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 10) | NC score | 0.886609 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 11) | NC score | 0.198388 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 7.59969e-07 (rank : 12) | NC score | 0.177194 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PP11_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 13) | NC score | 0.311499 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
VTNC_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.154892 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
SUSD2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.109766 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
TINAG_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.087795 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |
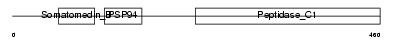
|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
VTNC_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.146440 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
NUCGL_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.067604 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2C4 | Gene names | ENDOGL1, ENGL | |||
|
Domain Architecture |
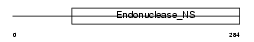
|
|||||
| Description | Endonuclease G-like 1 (EC 3.1.30.-) (Endo G-like). | |||||
|
EXTL3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.022289 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43909, O00225 | Gene names | EXTL3, EXTL1L, EXTR1, KIAA0519 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 3 (EC 2.4.1.223) (Glucuronyl-galactosyl-proteoglycan 4- alpha-N-acetylglucosaminyltransferase) (Putative tumor suppressor protein EXTL3) (Multiple exostosis-like protein 3) (Hereditary multiple exostoses gene isolog) (EXT-related protein 1). | |||||
|
SUSD2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.092404 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
PIGO_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.138932 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
SLIK5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.012871 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94991 | Gene names | SLITRK5, KIAA0918, LRRC11 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor (Leucine-rich repeat-containing protein 11). | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.033412 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.032781 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
SORL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.030090 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
SORL_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.029697 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
TENA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.025848 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
SLIK5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.011671 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q810B7, Q9CT21 | Gene names | Slitrk5 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor. | |||||
|
TENA_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.025820 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.027383 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
PIGO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.132008 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
CRUM1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.016813 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.016409 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
E2AK3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.002157 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZJ5, O95846 | Gene names | EIF2AK3, PEK, PERK | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 3 precursor (EC 2.7.11.1) (PRKR-like endoplasmic reticulum kinase) (Pancreatic eIF2-alpha kinase) (HsPEK). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.006252 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.025685 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.017603 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBLN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.014033 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
FBN1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.016433 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
LRP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.026541 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75096 | Gene names | LRP4, KIAA0816, MEGF7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (Multiple epidermal growth factor-like domains 7). | |||||
|
LRP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.026821 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
CP4F3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.002819 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08477, O60634, Q5U740 | Gene names | CYP4F3, LTB4H | |||
|
Domain Architecture |
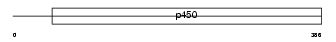
|
|||||
| Description | Cytochrome P450 4F3 (EC 1.14.13.30) (CYPIVF3) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.029226 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
ATRN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.010017 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
FAK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | -0.000300 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
FAK2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | -0.000495 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
GRN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.024797 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.055434 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
PIGG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.126907 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
PIGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.053391 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
PIGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.061919 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
ENPP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P22413, Q5T9R6, Q9NPZ3, Q9P1P6, Q9UP61, Q9Y6K3 | Gene names | ENPP1, NPPS, PC1, PDNP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP1_MOUSE
|
||||||
| NC score | 0.993516 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06802 | Gene names | Enpp1, Npps, Pc1, Pdnp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 1 (E-NPP 1) (Phosphodiesterase I/nucleotide pyrophosphatase 1) (Plasma-cell membrane glycoprotein PC-1) (Antigen Ly-41) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP3_HUMAN
|
||||||
| NC score | 0.989046 (rank : 3) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14638 | Gene names | ENPP3, PDNP3 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 3 (E-NPP 3) (Phosphodiesterase I/nucleotide pyrophosphatase 3) (Phosphodiesterase I beta) (PD-Ibeta) (CD203c antigen) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_MOUSE
|
||||||
| NC score | 0.975756 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9R1E6, Q99LG9 | Gene names | Enpp2, Npps2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP2_HUMAN
|
||||||
| NC score | 0.975312 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13822, Q13827, Q15117 | Gene names | ENPP2, ATX, PDNP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (E-NPP 2) (Phosphodiesterase I/nucleotide pyrophosphatase 2) (Phosphodiesterase I alpha) (PD-Ialpha) (Autotaxin) [Includes: Alkaline phosphodiesterase I (EC 3.1.4.1); Nucleotide pyrophosphatase (EC 3.6.1.9) (NPPase)]. | |||||
|
ENPP5_MOUSE
|
||||||
| NC score | 0.891055 (rank : 6) | θ value | 1.6097e-65 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EQG7, Q921P7 | Gene names | Enpp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP5_HUMAN
|
||||||
| NC score | 0.890968 (rank : 7) | θ value | 1.96772e-63 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UJA9, Q6UX49 | Gene names | ENPP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 5 precursor (EC 3.1.-.-) (E-NPP5) (NPP-5). | |||||
|
ENPP7_HUMAN
|
||||||
| NC score | 0.886609 (rank : 8) | θ value | 7.51208e-47 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWV6, Q6ZTS5, Q8IUS8 | Gene names | ENPP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 7 precursor (EC 3.1.4.12) (E-NPP7) (NPP-7) (Alkaline sphingomyelin phosphodiesterase) (Intestinal alkaline sphingomyelinase) (Alk-SMase). | |||||
|
ENPP6_HUMAN
|
||||||
| NC score | 0.868958 (rank : 9) | θ value | 6.56568e-51 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6UWR7, Q96M57 | Gene names | ENPP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
ENPP6_MOUSE
|
||||||
| NC score | 0.866532 (rank : 10) | θ value | 3.04986e-48 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BGN3, Q4VAH5 | Gene names | Enpp6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ectonucleotide pyrophosphatase/phosphodiesterase 6 precursor (EC 3.1.-.-) (E-NPP6) (NPP-6) [Contains: Ectonucleotide pyrophosphatase/phosphodiesterase 6 soluble form]. | |||||
|
PP11_HUMAN
|
||||||
| NC score | 0.311499 (rank : 11) | θ value | 9.29e-05 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P21128, Q2NKJ4 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Placental protein 11 precursor (EC 3.4.21.-) (PP11). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.198388 (rank : 12) | θ value | 3.08544e-08 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.177194 (rank : 13) | θ value | 7.59969e-07 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
VTNC_MOUSE
|
||||||
| NC score | 0.154892 (rank : 14) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P29788, Q8VII4 | Gene names | Vtn | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein). | |||||
|
VTNC_HUMAN
|
||||||
| NC score | 0.146440 (rank : 15) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P04004, P01141, Q9BSH7 | Gene names | VTN | |||
|
Domain Architecture |

|
|||||
| Description | Vitronectin precursor (Serum-spreading factor) (S-protein) (V75) [Contains: Vitronectin V65 subunit; Vitronectin V10 subunit; Somatomedin B]. | |||||
|
PIGO_HUMAN
|
||||||
| NC score | 0.138932 (rank : 16) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8TEQ8, Q6P154, Q6UX80, Q8TDS8, Q96CS9, Q9BVN9, Q9Y4B0 | Gene names | PIGO | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGO_MOUSE
|
||||||
| NC score | 0.132008 (rank : 17) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JJI6, Q9CRY2 | Gene names | Pigo | |||
|
Domain Architecture |

|
|||||
| Description | GPI ethanolamine phosphate transferase 3 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class O protein) (PIG-O). | |||||
|
PIGG_HUMAN
|
||||||
| NC score | 0.126907 (rank : 18) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5H8A4, Q2TAK5, Q6UX31, Q7L5Y4, Q8N866, Q8NCC9, Q96SY9, Q9BVT7, Q9NXG5 | Gene names | PIGG, GPI7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 2 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class G protein) (PIG-G) (GPI7 homolog) (hGPI7). | |||||
|
SUSD2_HUMAN
|
||||||
| NC score | 0.109766 (rank : 19) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UGT4, Q9H5Y6 | Gene names | SUSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
SUSD2_MOUSE
|
||||||
| NC score | 0.092404 (rank : 20) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9DBX3, Q8BY19, Q8BYN0, Q8BYZ3, Q8BYZ9, Q8BZQ4, Q8C179 | Gene names | Susd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 2 precursor. | |||||
|
TINAG_HUMAN
|
||||||
| NC score | 0.087795 (rank : 21) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UJW2, Q9UJW1, Q9ULZ4 | Gene names | TINAG | |||
|
Domain Architecture |

|
|||||
| Description | Tubulointerstitial nephritis antigen (TIN-Ag). | |||||
|
NUCGL_HUMAN
|
||||||
| NC score | 0.067604 (rank : 22) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2C4 | Gene names | ENDOGL1, ENGL | |||
|
Domain Architecture |

|
|||||
| Description | Endonuclease G-like 1 (EC 3.1.30.-) (Endo G-like). | |||||
|
PIGN_MOUSE
|
||||||
| NC score | 0.061919 (rank : 23) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R1S3, Q3V0S6, Q8VCC3, Q9R1S1, Q9R1S2 | Gene names | Pign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.055434 (rank : 24) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
PIGN_HUMAN
|
||||||
| NC score | 0.053391 (rank : 25) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95427, Q7L8F8, Q8TC01, Q9NT05 | Gene names | PIGN, MCD4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GPI ethanolamine phosphate transferase 1 (EC 2.-.-.-) (Phosphatidylinositol-glycan biosynthesis class N protein) (PIG-N) (MCD4 homolog). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.033412 (rank : 26) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.032781 (rank : 27) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
SORL_HUMAN
|
||||||
| NC score | 0.030090 (rank : 28) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92673, Q92856 | Gene names | SORL1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (SorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11). | |||||
|
SORL_MOUSE
|
||||||
| NC score | 0.029697 (rank : 29) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88307, O54711, O70581 | Gene names | Sorl1 | |||
|
Domain Architecture |

|
|||||
| Description | Sortilin-related receptor precursor (Sorting protein-related receptor containing LDLR class A repeats) (mSorLA) (SorLA-1) (Low-density lipoprotein receptor relative with 11 ligand-binding repeats) (LDLR relative with 11 ligand-binding repeats) (LR11) (Gp250). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.029226 (rank : 30) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
TENR_HUMAN
|
||||||
| NC score | 0.027383 (rank : 31) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 526 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92752, Q15568, Q5R3G0 | Gene names | TNR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin). | |||||
|
LRP4_MOUSE
|
||||||
| NC score | 0.026821 (rank : 32) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
LRP4_HUMAN
|
||||||
| NC score | 0.026541 (rank : 33) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75096 | Gene names | LRP4, KIAA0816, MEGF7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (Multiple epidermal growth factor-like domains 7). | |||||
|
TENA_MOUSE
|
||||||
| NC score | 0.025848 (rank : 34) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80YX1, Q64706, Q80YX2 | Gene names | Tnc, Hxb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion). | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.025820 (rank : 35) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.025685 (rank : 36) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
GRN_HUMAN
|
||||||
| NC score | 0.024797 (rank : 37) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28799, P23781, P23782, P23783, P23784, Q9BWE7, Q9UCH0 | Gene names | GRN | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) [Contains: Acrogranin; Paragranulin; Granulin-1 (Granulin G); Granulin-2 (Granulin F); Granulin-3 (Granulin B); Granulin-4 (Granulin A); Granulin-5 (Granulin C); Granulin-6 (Granulin D); Granulin-7 (Granulin E)]. | |||||
|
EXTL3_HUMAN
|
||||||
| NC score | 0.022289 (rank : 38) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43909, O00225 | Gene names | EXTL3, EXTL1L, EXTR1, KIAA0519 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 3 (EC 2.4.1.223) (Glucuronyl-galactosyl-proteoglycan 4- alpha-N-acetylglucosaminyltransferase) (Putative tumor suppressor protein EXTL3) (Multiple exostosis-like protein 3) (Hereditary multiple exostoses gene isolog) (EXT-related protein 1). | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.017603 (rank : 39) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
CRUM1_HUMAN
|
||||||
| NC score | 0.016813 (rank : 40) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P82279, Q5K3A6, Q5TC28, Q5VUT1, Q6N027, Q8WWY0 | Gene names | CRB1 | |||
|
Domain Architecture |

|
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.016433 (rank : 41) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.016409 (rank : 42) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
FBLN1_MOUSE
|
||||||
| NC score | 0.014033 (rank : 43) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
SLIK5_HUMAN
|
||||||
| NC score | 0.012871 (rank : 44) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94991 | Gene names | SLITRK5, KIAA0918, LRRC11 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor (Leucine-rich repeat-containing protein 11). | |||||
|
SLIK5_MOUSE
|
||||||
| NC score | 0.011671 (rank : 45) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q810B7, Q9CT21 | Gene names | Slitrk5 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT and NTRK-like protein 5 precursor. | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.010017 (rank : 46) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.006252 (rank : 47) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
CP4F3_HUMAN
|
||||||
| NC score | 0.002819 (rank : 48) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08477, O60634, Q5U740 | Gene names | CYP4F3, LTB4H | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 4F3 (EC 1.14.13.30) (CYPIVF3) (Leukotriene-B(4) omega- hydroxylase) (Leukotriene-B(4) 20-monooxygenase) (Cytochrome P450-LTB- omega). | |||||
|
E2AK3_HUMAN
|
||||||
| NC score | 0.002157 (rank : 49) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZJ5, O95846 | Gene names | EIF2AK3, PEK, PERK | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 2-alpha kinase 3 precursor (EC 2.7.11.1) (PRKR-like endoplasmic reticulum kinase) (Pancreatic eIF2-alpha kinase) (HsPEK). | |||||
|
FAK2_HUMAN
|
||||||
| NC score | -0.000300 (rank : 50) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
FAK2_MOUSE
|
||||||
| NC score | -0.000495 (rank : 51) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||