Please be patient as the page loads
|
SMAD1_HUMAN
|
||||||
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMAD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999236 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.994862 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993660 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992661 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD9_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.988309 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| θ value | 5.9766e-145 (rank : 7) | NC score | 0.984398 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| θ value | 5.9766e-145 (rank : 8) | NC score | 0.984398 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| θ value | 1.16911e-132 (rank : 9) | NC score | 0.981136 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| θ value | 1.16911e-132 (rank : 10) | NC score | 0.981138 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 4.23606e-66 (rank : 11) | NC score | 0.922510 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 2.74572e-65 (rank : 12) | NC score | 0.923974 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD6_MOUSE
|
||||||
| θ value | 2.27762e-19 (rank : 13) | NC score | 0.768487 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 14) | NC score | 0.765554 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 15) | NC score | 0.751467 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
SMAD7_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 16) | NC score | 0.752282 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
IRF4_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 17) | NC score | 0.060773 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.046055 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.016832 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.096501 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DAZ1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.060877 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.061411 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
DAZ3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 23) | NC score | 0.061606 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
DAZ4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 24) | NC score | 0.060939 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 25) | NC score | 0.056798 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CD248_MOUSE
|
||||||
| θ value | 0.125558 (rank : 26) | NC score | 0.020018 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.024898 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.010022 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.023024 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
EFS_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.019174 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.009831 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.015034 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.009914 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.016154 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.014681 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.022104 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.009430 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
NDF1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.009245 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
PO3F4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.006552 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49335, Q5H9G9, Q99410 | Gene names | POU3F4, BRN4, OTF9 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
RUNX3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.009175 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
LCP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.043430 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
NPAS4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.016635 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.011537 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
P73_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.017938 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.009128 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.013090 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
GT2D1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.022476 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
IRF7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.025303 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
KBTB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.015556 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
NDF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.007256 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
ZEP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | -0.001170 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||
|
ZN645_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.018069 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.002974 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MEF2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.009502 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015493 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.015552 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
SPR1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.003782 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
SYT9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.001003 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |
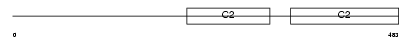
|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
WNT16_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.005318 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.999236 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.994862 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.993660 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.992661 (rank : 5) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD9_MOUSE
|
||||||
| NC score | 0.988309 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD3_HUMAN
|
||||||
| NC score | 0.984398 (rank : 7) | θ value | 5.9766e-145 (rank : 7) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| NC score | 0.984398 (rank : 8) | θ value | 5.9766e-145 (rank : 8) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_MOUSE
|
||||||
| NC score | 0.981138 (rank : 9) | θ value | 1.16911e-132 (rank : 10) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD2_HUMAN
|
||||||
| NC score | 0.981136 (rank : 10) | θ value | 1.16911e-132 (rank : 9) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.923974 (rank : 11) | θ value | 2.74572e-65 (rank : 12) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.922510 (rank : 12) | θ value | 4.23606e-66 (rank : 11) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.768487 (rank : 13) | θ value | 2.27762e-19 (rank : 13) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| NC score | 0.765554 (rank : 14) | θ value | 1.47631e-18 (rank : 14) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_MOUSE
|
||||||
| NC score | 0.752282 (rank : 15) | θ value | 3.07829e-16 (rank : 16) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD7_HUMAN
|
||||||
| NC score | 0.751467 (rank : 16) | θ value | 3.07829e-16 (rank : 15) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.096501 (rank : 17) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DAZ3_HUMAN
|
||||||
| NC score | 0.061606 (rank : 18) | θ value | 0.0961366 (rank : 23) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
DAZ2_HUMAN
|
||||||
| NC score | 0.061411 (rank : 19) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
DAZ4_HUMAN
|
||||||
| NC score | 0.060939 (rank : 20) | θ value | 0.0961366 (rank : 24) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
DAZ1_HUMAN
|
||||||
| NC score | 0.060877 (rank : 21) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
IRF4_HUMAN
|
||||||
| NC score | 0.060773 (rank : 22) | θ value | 0.000270298 (rank : 17) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15306, Q99660 | Gene names | IRF4, MUM1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 4 (IRF-4) (Lymphocyte-specific interferon regulatory factor) (LSIRF) (NF-EM5) (Multiple myeloma oncogene 1). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.056798 (rank : 23) | θ value | 0.0961366 (rank : 25) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.046055 (rank : 24) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
LCP2_HUMAN
|
||||||
| NC score | 0.043430 (rank : 25) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13094 | Gene names | LCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte cytosolic protein 2 (SH2 domain-containing leukocyte protein of 76 kDa) (SLP-76 tyrosine phosphoprotein) (SLP76). | |||||
|
IRF7_HUMAN
|
||||||
| NC score | 0.025303 (rank : 26) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92985, O00331, O00332, O00333, O75924 | Gene names | IRF7 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 7 (IRF-7). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.024898 (rank : 27) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.023024 (rank : 28) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
GT2D1_MOUSE
|
||||||
| NC score | 0.022476 (rank : 29) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JI57, Q8VHD5, Q8VI58, Q9EQE7, Q9ESZ6, Q9ESZ7 | Gene names | Gtf2ird1, Ben | |||
|
Domain Architecture |

|
|||||
| Description | General transcription factor II-I repeat domain-containing protein 1 (GTF2I repeat domain-containing protein 1) (Binding factor for early enhancer). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.022104 (rank : 30) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
CD248_MOUSE
|
||||||
| NC score | 0.020018 (rank : 31) | θ value | 0.125558 (rank : 26) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91V98, Q3UMV6, Q91ZV1 | Gene names | Cd248, Tem1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.019174 (rank : 32) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
ZN645_HUMAN
|
||||||
| NC score | 0.018069 (rank : 33) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N7E2, Q6DJY9 | Gene names | ZNF645 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 645. | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.017938 (rank : 34) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.016832 (rank : 35) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
NPAS4_HUMAN
|
||||||
| NC score | 0.016635 (rank : 36) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IUM7, Q8N8S5, Q8N9Q9 | Gene names | NPAS4, NXF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuronal PAS domain-containing protein 4 (Neuronal PAS4) (HLH-PAS transcription factor NXF). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.016154 (rank : 37) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
KBTB4_HUMAN
|
||||||
| NC score | 0.015556 (rank : 38) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.015552 (rank : 39) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.015493 (rank : 40) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.015034 (rank : 41) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.014681 (rank : 42) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.013090 (rank : 43) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.011537 (rank : 44) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.010022 (rank : 45) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.009914 (rank : 46) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.009831 (rank : 47) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
MEF2B_HUMAN
|
||||||
| NC score | 0.009502 (rank : 48) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02080 | Gene names | MEF2B, XMEF2 | |||
|
Domain Architecture |

|
|||||
| Description | Myocyte-specific enhancer factor 2B (Serum response factor-like protein 2) (XMEF2) (RSRFR2). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.009430 (rank : 49) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
NDF1_MOUSE
|
||||||
| NC score | 0.009245 (rank : 50) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60867, Q545N9, Q60897 | Gene names | Neurod1, Neurod | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1). | |||||
|
RUNX3_HUMAN
|
||||||
| NC score | 0.009175 (rank : 51) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.009128 (rank : 52) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
NDF1_HUMAN
|
||||||
| NC score | 0.007256 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13562, O00343, Q13340, Q5U095, Q96TH0, Q99455, Q9UEC8 | Gene names | NEUROD1, NEUROD | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic differentiation factor 1 (NeuroD1) (NeuroD). | |||||
|
PO3F4_HUMAN
|
||||||
| NC score | 0.006552 (rank : 54) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49335, Q5H9G9, Q99410 | Gene names | POU3F4, BRN4, OTF9 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 4 (Brain-specific homeobox/POU domain protein 4) (Brain-4) (Brn-4). | |||||
|
WNT16_MOUSE
|
||||||
| NC score | 0.005318 (rank : 55) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QYS1 | Gene names | Wnt16 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Wnt-16 precursor. | |||||
|
SPR1B_MOUSE
|
||||||
| NC score | 0.003782 (rank : 56) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62267 | Gene names | Sprr1b | |||
|
Domain Architecture |

|
|||||
| Description | Cornifin B (Small proline-rich protein 1B) (SPR1B) (SPR1 B). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.002974 (rank : 57) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
SYT9_MOUSE
|
||||||
| NC score | 0.001003 (rank : 58) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R0N9, Q8C280 | Gene names | Syt9, Syt5 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-9 (Synaptotagmin IX) (SytIX) (Synaptotagmin V). | |||||
|
ZEP2_MOUSE
|
||||||
| NC score | -0.001170 (rank : 59) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 59 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UHF7, O55140, Q3UVD4, Q3UVH5 | Gene names | Hivep2, Mibp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Human immunodeficiency virus type I enhancer-binding protein 2 homolog (Myc intron-binding protein 1) (MIBP-1). | |||||