Please be patient as the page loads
|
SMAD6_HUMAN
|
||||||
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMAD6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997222 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD7_HUMAN
|
||||||
| θ value | 1.54831e-84 (rank : 3) | NC score | 0.971779 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
SMAD7_MOUSE
|
||||||
| θ value | 2.91998e-83 (rank : 4) | NC score | 0.971790 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD3_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 5) | NC score | 0.812134 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |
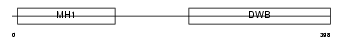
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| θ value | 1.85889e-29 (rank : 6) | NC score | 0.812134 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |
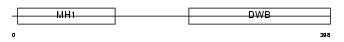
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 7) | NC score | 0.810315 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| θ value | 1.2049e-28 (rank : 8) | NC score | 0.810357 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD9_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 9) | NC score | 0.799259 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |
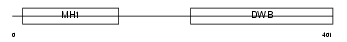
|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD4_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 10) | NC score | 0.782268 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 1.2077e-20 (rank : 11) | NC score | 0.777164 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 12) | NC score | 0.772192 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 13) | NC score | 0.765501 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD1_HUMAN
|
||||||
| θ value | 1.47631e-18 (rank : 14) | NC score | 0.765554 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 1.47631e-18 (rank : 15) | NC score | 0.763852 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 2.5182e-18 (rank : 16) | NC score | 0.764605 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.006400 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
DCC_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.009491 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.013036 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.008283 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.030806 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.015752 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
IBP6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.014241 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |
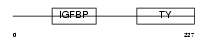
|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.010128 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
DCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.007992 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
HIC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | -0.001560 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |
|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
PAK4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | -0.001773 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||
|
ASPM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.008284 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.010004 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.006226 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.004159 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
SMAD6_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.997222 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD7_MOUSE
|
||||||
| NC score | 0.971790 (rank : 3) | θ value | 2.91998e-83 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD7_HUMAN
|
||||||
| NC score | 0.971779 (rank : 4) | θ value | 1.54831e-84 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
SMAD3_HUMAN
|
||||||
| NC score | 0.812134 (rank : 5) | θ value | 1.85889e-29 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| NC score | 0.812134 (rank : 6) | θ value | 1.85889e-29 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD2_MOUSE
|
||||||
| NC score | 0.810357 (rank : 7) | θ value | 1.2049e-28 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD2_HUMAN
|
||||||
| NC score | 0.810315 (rank : 8) | θ value | 1.2049e-28 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD9_MOUSE
|
||||||
| NC score | 0.799259 (rank : 9) | θ value | 2.05525e-28 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.782268 (rank : 10) | θ value | 1.2077e-20 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.777164 (rank : 11) | θ value | 1.2077e-20 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.772192 (rank : 12) | θ value | 2.97466e-19 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.765554 (rank : 13) | θ value | 1.47631e-18 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.765501 (rank : 14) | θ value | 8.65492e-19 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.764605 (rank : 15) | θ value | 2.5182e-18 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.763852 (rank : 16) | θ value | 1.47631e-18 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.030806 (rank : 17) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.015752 (rank : 18) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
IBP6_MOUSE
|
||||||
| NC score | 0.014241 (rank : 19) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P47880 | Gene names | Igfbp6, Igfbp-6 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor-binding protein 6 precursor (IGFBP-6) (IBP- 6) (IGF-binding protein 6). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.013036 (rank : 20) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.010128 (rank : 21) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.010004 (rank : 22) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
DCC_MOUSE
|
||||||
| NC score | 0.009491 (rank : 23) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70211 | Gene names | Dcc | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC). | |||||
|
ASPM_MOUSE
|
||||||
| NC score | 0.008284 (rank : 24) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CJ27, O88482, Q8BJI8, Q8BKT4 | Gene names | Aspm, Calmbp1, Sha1 | |||
|
Domain Architecture |

|
|||||
| Description | Abnormal spindle-like microcephaly-associated protein homolog (Calmodulin-binding protein 1) (Spindle and hydroxyurea checkpoint abnormal protein) (Calmodulin-binding protein Sha1). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.008283 (rank : 25) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
DCC_HUMAN
|
||||||
| NC score | 0.007992 (rank : 26) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 583 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43146 | Gene names | DCC | |||
|
Domain Architecture |

|
|||||
| Description | Netrin receptor DCC precursor (Tumor suppressor protein DCC) (Colorectal cancer suppressor). | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.006400 (rank : 27) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.006226 (rank : 28) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.004159 (rank : 29) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
HIC1_MOUSE
|
||||||
| NC score | -0.001560 (rank : 30) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 863 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1Y5, Q9R1Y6, Q9R2B0 | Gene names | Hic1 | |||
|
Domain Architecture |

|
|||||
| Description | Hypermethylated in cancer 1 protein (Hic-1). | |||||
|
PAK4_MOUSE
|
||||||
| NC score | -0.001773 (rank : 31) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1098 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTW9, Q6ZPX0, Q80Z97, Q9CS71 | Gene names | Pak4, Kiaa1142 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase PAK 4 (EC 2.7.11.1) (p21-activated kinase 4) (PAK-4). | |||||