Please be patient as the page loads
|
SMAD4_HUMAN
|
||||||
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMAD4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985217 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD3_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 3) | NC score | 0.929801 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| θ value | 1.90147e-66 (rank : 4) | NC score | 0.929801 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD9_HUMAN
|
||||||
| θ value | 2.4834e-66 (rank : 5) | NC score | 0.926339 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD5_HUMAN
|
||||||
| θ value | 7.2256e-66 (rank : 6) | NC score | 0.925775 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| θ value | 7.2256e-66 (rank : 7) | NC score | 0.924655 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD1_HUMAN
|
||||||
| θ value | 2.74572e-65 (rank : 8) | NC score | 0.923974 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| θ value | 3.96481e-64 (rank : 9) | NC score | 0.923628 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD2_HUMAN
|
||||||
| θ value | 1.2411e-41 (rank : 10) | NC score | 0.909903 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD2_MOUSE
|
||||||
| θ value | 1.2411e-41 (rank : 11) | NC score | 0.909937 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD9_MOUSE
|
||||||
| θ value | 4.88048e-38 (rank : 12) | NC score | 0.915432 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD6_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 13) | NC score | 0.782268 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
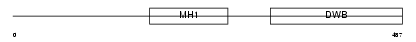
SMAD6_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 14) | NC score | 0.783315 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
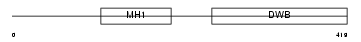
SMAD7_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 15) | NC score | 0.766683 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
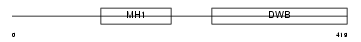
SMAD7_MOUSE
|
||||||
| θ value | 2.7842e-17 (rank : 16) | NC score | 0.767538 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |
|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.017940 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
RNF12_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.026246 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.054697 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ARAF_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.001982 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.043591 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.042371 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.053445 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.052928 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.052617 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.027336 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.052310 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PODXL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.043561 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.052121 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.001939 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
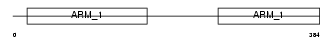
ADRM1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.044041 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |
|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
BORG5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.047740 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |
|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
FOXJ3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.023922 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.051638 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GLP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.053890 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14220 | Gene names | Gypa | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin. | |||||
|
LRC41_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.030988 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15345, Q71RA8, Q9BSM0 | Gene names | LRRC41, MUF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.024284 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.002698 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.051942 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.052188 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ADRM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.033034 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.010112 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
NCOA5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.029267 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.011441 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.001166 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
NCOA5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.027024 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
UB7I3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.012827 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |
|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
FOXN4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.007434 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Q3, Q920C0 | Gene names | Foxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N4. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.051089 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PA24B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.009865 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.006529 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PORIM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.074349 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N131, Q8IWS2, Q96QV2 | Gene names | TMEM123, KCT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123) (Pro-oncosis receptor inducing membrane injury) (Keratinocytes-associated transmembrane protein 3) (KCT-3). | |||||
|
X3CL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.014223 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |
|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
DLG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.004859 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.009576 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.017485 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CCD22_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.007155 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIG7, Q8BYH4 | Gene names | Ccdc22, DXImx40e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 22. | |||||
|
DACH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.012692 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.012896 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.029284 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
ZF106_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.004625 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.007310 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.008490 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.015585 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
CNTN3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.000074 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.001782 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.006243 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NPAS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.003852 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
RCOR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.010729 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.011631 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.011605 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.006579 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.060284 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
SMAD4_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q13485 | Gene names | SMAD4, DPC4, MADH4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4) (hSMAD4). | |||||
|
SMAD4_MOUSE
|
||||||
| NC score | 0.985217 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97471, Q9CW56 | Gene names | Smad4, Dpc4, Madh4 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 4 (SMAD 4) (Mothers against DPP homolog 4) (Deletion target in pancreatic carcinoma 4 homolog) (Smad4). | |||||
|
SMAD3_HUMAN
|
||||||
| NC score | 0.929801 (rank : 3) | θ value | 1.90147e-66 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84022, O09064, O09144, O14510, O35273, Q92940, Q93002, Q9GKR4 | Gene names | SMAD3, MADH3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (hMAD-3) (JV15-2) (hSMAD3). | |||||
|
SMAD3_MOUSE
|
||||||
| NC score | 0.929801 (rank : 4) | θ value | 1.90147e-66 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BUN5, O09064, O09144, O14510, O35273, Q8BX84, Q92940, Q93002, Q9GKR4 | Gene names | Smad3, Madh3 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 3 (SMAD 3) (Mothers against DPP homolog 3) (Mad3) (mMad3). | |||||
|
SMAD9_HUMAN
|
||||||
| NC score | 0.926339 (rank : 5) | θ value | 2.4834e-66 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15198, O14989 | Gene names | SMAD9, MADH6, MADH9 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Madh6). | |||||
|
SMAD5_HUMAN
|
||||||
| NC score | 0.925775 (rank : 6) | θ value | 7.2256e-66 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99717, O14688, Q15798, Q9UQA1 | Gene names | SMAD5, MADH5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (hSmad5) (JV5-1). | |||||
|
SMAD5_MOUSE
|
||||||
| NC score | 0.924655 (rank : 7) | θ value | 7.2256e-66 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97454, P70341, Q810K0 | Gene names | Smad5, Madh5, Msmad5 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 5 (SMAD 5) (Mothers against DPP homolog 5) (Smad5) (mSmad5) (Dwarfin-C) (Dwf-C). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.923974 (rank : 8) | θ value | 2.74572e-65 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SMAD1_MOUSE
|
||||||
| NC score | 0.923628 (rank : 9) | θ value | 3.96481e-64 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P70340, P70442, Q9CYK6 | Gene names | Smad1, Madh1, Madr1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Mothers-against-DPP-related 1) (mMad1) (Dwarfin-A) (Dwf-A). | |||||
|
SMAD9_MOUSE
|
||||||
| NC score | 0.915432 (rank : 10) | θ value | 4.88048e-38 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIW5, Q8CFF9 | Gene names | Smad9, Madh8, Madh9, Smad8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 9 (SMAD 9) (Mothers against DPP homolog 9) (Smad9) (Smad8). | |||||
|
SMAD2_MOUSE
|
||||||
| NC score | 0.909937 (rank : 11) | θ value | 1.2411e-41 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62432, Q9D8P6 | Gene names | Smad2, Madh2, Madr2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (mMad2). | |||||
|
SMAD2_HUMAN
|
||||||
| NC score | 0.909903 (rank : 12) | θ value | 1.2411e-41 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15796 | Gene names | SMAD2, MADH2, MADR2 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 2 (SMAD 2) (Mothers against DPP homolog 2) (Mad-related protein 2) (hMAD-2) (JV18-1) (hSMAD2). | |||||
|
SMAD6_MOUSE
|
||||||
| NC score | 0.783315 (rank : 13) | θ value | 2.06002e-20 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35182, Q9CW62 | Gene names | Smad6, Madh6, Madh7, Msmad6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (Mad homolog 7). | |||||
|
SMAD6_HUMAN
|
||||||
| NC score | 0.782268 (rank : 14) | θ value | 1.2077e-20 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O43541, O43654, Q15799, Q9UKZ3 | Gene names | SMAD6, MADH6 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 6 (SMAD 6) (Mothers against DPP homolog 6) (Smad6) (hSMAD6). | |||||
|
SMAD7_MOUSE
|
||||||
| NC score | 0.767538 (rank : 15) | θ value | 2.7842e-17 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O35253, O88709 | Gene names | Smad7, Madh7 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (Madh8). | |||||
|
SMAD7_HUMAN
|
||||||
| NC score | 0.766683 (rank : 16) | θ value | 2.7842e-17 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15105, O14740, Q6DK23 | Gene names | SMAD7, MADH7, MADH8 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 7 (SMAD 7) (Mothers against DPP homolog 7) (Smad7) (hSMAD7). | |||||
|
PORIM_HUMAN
|
||||||
| NC score | 0.074349 (rank : 17) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N131, Q8IWS2, Q96QV2 | Gene names | TMEM123, KCT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Porimin precursor (Transmembrane protein 123) (Pro-oncosis receptor inducing membrane injury) (Keratinocytes-associated transmembrane protein 3) (KCT-3). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.060284 (rank : 18) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.054697 (rank : 19) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GLP_MOUSE
|
||||||
| NC score | 0.053890 (rank : 20) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P14220 | Gene names | Gypa | |||
|
Domain Architecture |

|
|||||
| Description | Glycophorin. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.053445 (rank : 21) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.052928 (rank : 22) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.052617 (rank : 23) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.052310 (rank : 24) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.052188 (rank : 25) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.052121 (rank : 26) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.051942 (rank : 27) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.051638 (rank : 28) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.051089 (rank : 29) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
BORG5_HUMAN
|
||||||
| NC score | 0.047740 (rank : 30) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00587, Q96GN1 | Gene names | CDC42EP1, BORG5, CEP1, MSE55 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42 effector protein 1 (Binder of Rho GTPases 5) (Serum protein MSE55). | |||||
|
ADRM1_HUMAN
|
||||||
| NC score | 0.044041 (rank : 31) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16186, Q9H1P2 | Gene names | ADRM1, GP110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.043591 (rank : 32) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
PODXL_MOUSE
|
||||||
| NC score | 0.043561 (rank : 33) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R0M4, Q9ESZ1 | Gene names | Podxl, Pclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.042371 (rank : 34) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ADRM1_MOUSE
|
||||||
| NC score | 0.033034 (rank : 35) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JKV1 | Gene names | Adrm1, Gp110 | |||
|
Domain Architecture |

|
|||||
| Description | Adhesion-regulating molecule 1 precursor (110 kDa cell membrane glycoprotein) (Gp110) (ARM-1). | |||||
|
LRC41_HUMAN
|
||||||
| NC score | 0.030988 (rank : 36) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15345, Q71RA8, Q9BSM0 | Gene names | LRRC41, MUF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.029284 (rank : 37) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NCOA5_HUMAN
|
||||||
| NC score | 0.029267 (rank : 38) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.027336 (rank : 39) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
NCOA5_MOUSE
|
||||||
| NC score | 0.027024 (rank : 40) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91W39 | Gene names | Ncoa5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
RNF12_HUMAN
|
||||||
| NC score | 0.026246 (rank : 41) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NVW2, Q9Y598 | Gene names | RNF12, RLIM | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM) (NY-REN-43 antigen). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.024284 (rank : 42) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
FOXJ3_HUMAN
|
||||||
| NC score | 0.023922 (rank : 43) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPW0, Q9NSS7 | Gene names | FOXJ3, KIAA1041 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J3. | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.017940 (rank : 44) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.017485 (rank : 45) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.015585 (rank : 46) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
X3CL1_MOUSE
|
||||||
| NC score | 0.014223 (rank : 47) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35188, O35933 | Gene names | Cx3cl1, Cx3c, Fkn, Scyd1 | |||
|
Domain Architecture |

|
|||||
| Description | Fractalkine precursor (CX3CL1) (Neurotactin) (CX3C membrane-anchored chemokine) (Small inducible cytokine D1). | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.012896 (rank : 48) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
UB7I3_MOUSE
|
||||||
| NC score | 0.012827 (rank : 49) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUB0 | Gene names | Rbck1, Rbck, Ubce7ip3, Uip28 | |||
|
Domain Architecture |

|
|||||
| Description | RanBP-type and C3HC4-type zinc finger-containing protein 1 (Ubiquitin- conjugating enzyme 7-interacting protein 3) (UbcM4-interacting protein 28). | |||||
|
DACH1_HUMAN
|
||||||
| NC score | 0.012692 (rank : 50) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI36, O75523, O75687, Q5VYY3, Q5VYY4, Q96SG3, Q96SG4, Q9H524, Q9UMH4 | Gene names | DACH1, DACH | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.011631 (rank : 51) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.011605 (rank : 52) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.011441 (rank : 53) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
RCOR3_HUMAN
|
||||||
| NC score | 0.010729 (rank : 54) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P2K3, Q5VT47, Q7L9I5, Q8N5U3, Q9NV83 | Gene names | RCOR3, KIAA1343 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | REST corepressor 3. | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.010112 (rank : 55) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
PA24B_HUMAN
|
||||||
| NC score | 0.009865 (rank : 56) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95712, Q59GF9, Q8TB10, Q9UKV7 | Gene names | PLA2G4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.009576 (rank : 57) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.008490 (rank : 58) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
FOXN4_MOUSE
|
||||||
| NC score | 0.007434 (rank : 59) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Q3, Q920C0 | Gene names | Foxn4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N4. | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.007310 (rank : 60) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
CCD22_MOUSE
|
||||||
| NC score | 0.007155 (rank : 61) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIG7, Q8BYH4 | Gene names | Ccdc22, DXImx40e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 22. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.006579 (rank : 62) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.006529 (rank : 63) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.006243 (rank : 64) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
DLG2_HUMAN
|
||||||
| NC score | 0.004859 (rank : 65) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15700, Q59G57, Q68CQ8, Q6ZTA8 | Gene names | DLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 2 (Postsynaptic density protein PSD-93) (Channel- associated protein of synapse-110) (Chapsyn-110). | |||||
|
ZF106_MOUSE
|
||||||
| NC score | 0.004625 (rank : 66) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88466, O55185, O88465, O88467, Q792M1, Q792P4, Q8CDZ8, Q8R3I4, Q9ESU3 | Gene names | Zfp106, H3a, Sh3bp3, Sirm, Znf474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 (Zfp-106) (Zinc finger protein 474) (H3a minor histocompatibility antigen) (Son of insulin receptor mutant). | |||||
|
NPAS1_HUMAN
|
||||||
| NC score | 0.003852 (rank : 67) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99742, Q99632, Q9BY83 | Gene names | NPAS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal PAS domain-containing protein 1 (Neuronal PAS1) (Member of PAS protein 5) (MOP5). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.002698 (rank : 68) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
ARAF_MOUSE
|
||||||
| NC score | 0.001982 (rank : 69) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04627, Q99J44, Q9CTT5, Q9D6R6, Q9DBU7 | Gene names | Araf, A-raf, Araf1 | |||
|
Domain Architecture |

|
|||||
| Description | A-Raf proto-oncogene serine/threonine-protein kinase (EC 2.7.11.1). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.001939 (rank : 70) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.001782 (rank : 71) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.001166 (rank : 72) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
CNTN3_HUMAN
|
||||||
| NC score | 0.000074 (rank : 73) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P232, Q9H039 | Gene names | CNTN3, KIAA1496, PANG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-3 precursor (Brain-derived immunoglobulin superfamily protein 1) (BIG-1) (Plasmacytoma-associated neuronal glycoprotein). | |||||