Please be patient as the page loads
|
GFC1_HUMAN
|
||||||
| SwissProt Accessions | P63117 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-F(c)1_Xq21.33 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GFC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P63117 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-F(c)1_Xq21.33 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein]. | |||||
|
GAW2_HUMAN
|
||||||
| θ value | 1.2784e-54 (rank : 2) | NC score | 0.881723 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRZ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-W_3q26.32 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein (Gag P15); Capsid protein (Core shell protein) (Gag P30)]. | |||||
|
GAH2_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 3) | NC score | 0.851818 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63118 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_3q26 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAH6_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 4) | NC score | 0.848714 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8A4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_22q11.2 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 5) | NC score | 0.019072 (rank : 111) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.072733 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.067681 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.023312 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.039672 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.063168 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.041317 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.015298 (rank : 122) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
ZSCA5_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.008883 (rank : 130) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
MYO15_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.026422 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.051038 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.056037 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.038948 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.043723 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.055061 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.044746 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.051645 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.013165 (rank : 126) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.013953 (rank : 124) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.024073 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
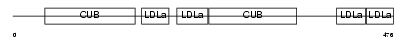
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.056286 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RECQ4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.028898 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.024537 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.039474 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.014964 (rank : 123) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
HXD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.016473 (rank : 118) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01822 | Gene names | Hoxd1, Hox-4.9, Hoxd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1 (Hox-4.9). | |||||
|
RBM4B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.042331 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.042275 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.042542 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.042476 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
SOX5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.026426 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
SOX5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.026233 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
COIA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.035902 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
HXD1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.015643 (rank : 120) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZZ0 | Gene names | HOXD1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1. | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.023181 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.065499 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
NEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.007464 (rank : 133) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.041718 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.018122 (rank : 114) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.018141 (rank : 113) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.017385 (rank : 116) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
CPSF4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.028105 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.053351 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.022747 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.042825 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.062880 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.065098 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SPAG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.015326 (rank : 121) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
ZN409_HUMAN
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.022941 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
ZN688_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.004865 (rank : 135) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
AOC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.021825 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75106, O00120, O75105, Q4TTW5, Q9UNY0 | Gene names | AOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retina-specific copper amine oxidase precursor (EC 1.4.3.6) (RAO) (Amine oxidase [copper-containing]). | |||||
|
AOC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.021825 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16853, Q45F94 | Gene names | AOC3, VAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane copper amine oxidase (EC 1.4.3.6) (Semicarbazide-sensitive amine oxidase) (SSAO) (Vascular adhesion protein 1) (VAP-1) (HPAO). | |||||
|
BI2L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.017997 (rank : 115) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHR4, Q75L21, Q75L22, Q96CV4, Q9Y2M8 | Gene names | BAIAP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAI1-associated protein 2-like protein 1). | |||||
|
CN145_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.009723 (rank : 129) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CSPG5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.026870 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
FAIM2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.020167 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BWQ8, Q9UJY9, Q9Y2F7 | Gene names | FAIM2, KIAA0950, LFG, TMBIM2 | |||
|
Domain Architecture |
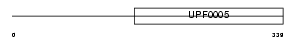
|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein) (Transmembrane BAX inhibitor motif-containing protein 2). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.007727 (rank : 132) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
LMBL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.024714 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.028286 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.044556 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.043002 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SCAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.019251 (rank : 109) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.026251 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.031085 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CD2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.025377 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
HXB13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.009992 (rank : 128) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92826, Q96QM4, Q99810 | Gene names | HOXB13 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B13. | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.039718 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
NUMBL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.020643 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
PER3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.024760 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.019101 (rank : 110) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
RIN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.046138 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.036570 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.028672 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
ZN384_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.003358 (rank : 137) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TF68, O15407, Q8N938 | Gene names | ZNF384, CAGH1, CIZ, NMP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 384 (Nuclear matrix transcription factor 4) (Nuclear matrix protein 4) (CAG repeat protein 1) (CAS-interacting zinc finger protein). | |||||
|
ANR11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.018246 (rank : 112) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.019365 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.058698 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.013716 (rank : 125) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
ERF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.015698 (rank : 119) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
ERF_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.016992 (rank : 117) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
GP152_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.010515 (rank : 127) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.025689 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MOES_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.004899 (rank : 134) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.003451 (rank : 136) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
OGFR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.033190 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
PAX9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.008254 (rank : 131) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |
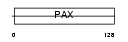
|
|||||
| Description | Paired box protein Pax-9. | |||||
|
PMYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.003184 (rank : 138) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESG9, Q8R3L4 | Gene names | Pkmyt1, Myt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.031763 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
RASF7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.019703 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |
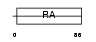
|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.044362 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
ZN395_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.024928 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8N7, Q6F6H2, Q9BY72, Q9NPB2, Q9NS57, Q9NS58, Q9NS59 | Gene names | ZNF395, HDBP2, PBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 395 (Papillomavirus-binding factor) (Papillomavirus regulatory factor 1) (PRF-1) (Huntington disease gene regulatory region-binding protein 2) (HDBP-2) (HD gene regulatory region-binding protein 2) (HD-regulating factor 2) (HDRF-2). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.053796 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.058741 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.063941 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.063594 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050848 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.053789 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054895 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CS016_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052144 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053281 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
FMNL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050485 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051769 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GGN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.056043 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.066657 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.061187 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.073069 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.051701 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.064448 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.068482 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052773 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.077178 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.093280 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.052463 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.055640 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.073834 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.063812 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063593 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.058042 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053328 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052902 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.066339 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.056213 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.053305 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.053996 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.066724 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.067215 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.053163 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.056368 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.059731 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.055342 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.059421 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.053664 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.065591 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.050963 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
GFC1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P63117 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-F(c)1_Xq21.33 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein]. | |||||
|
GAW2_HUMAN
|
||||||
| NC score | 0.881723 (rank : 2) | θ value | 1.2784e-54 (rank : 2) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRZ4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-W_3q26.32 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein (Gag P15); Capsid protein (Core shell protein) (Gag P30)]. | |||||
|
GAH2_HUMAN
|
||||||
| NC score | 0.851818 (rank : 3) | θ value | 2.96089e-35 (rank : 3) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63118 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_3q26 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAH6_HUMAN
|
||||||
| NC score | 0.848714 (rank : 4) | θ value | 3.61944e-33 (rank : 4) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8A4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-H_22q11.2 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.093280 (rank : 5) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.077178 (rank : 6) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.073834 (rank : 7) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.073069 (rank : 8) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.072733 (rank : 9) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.068482 (rank : 10) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.067681 (rank : 11) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.067215 (rank : 12) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.066724 (rank : 13) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.066657 (rank : 14) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.066339 (rank : 15) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.065591 (rank : 16) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.065499 (rank : 17) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.065098 (rank : 18) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.064448 (rank : 19) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.063941 (rank : 20) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.063812 (rank : 21) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.063594 (rank : 22) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.063593 (rank : 23) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.063168 (rank : 24) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.062880 (rank : 25) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.061187 (rank : 26) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.059731 (rank : 27) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.059421 (rank : 28) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.058741 (rank : 29) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.058698 (rank : 30) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.058042 (rank : 31) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.056368 (rank : 32) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.056286 (rank : 33) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.056213 (rank : 34) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.056043 (rank : 35) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.056037 (rank : 36) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.055640 (rank : 37) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.055342 (rank : 38) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.055061 (rank : 39) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.054895 (rank : 40) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
WASF2_MOUSE
|
||||||
| NC score | 0.053996 (rank : 41) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 315 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BH43 | Gene names | Wasf2, Wave2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 2 (WASP-family protein member 2) (Protein WAVE-2). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.053796 (rank : 42) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.053789 (rank : 43) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.053664 (rank : 44) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.053351 (rank : 45) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.053328 (rank : 46) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.053305 (rank : 47) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.053281 (rank : 48) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.053163 (rank : 49) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.052902 (rank : 50) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.052773 (rank : 51) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.052463 (rank : 52) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
CS016_HUMAN
|
||||||
| NC score | 0.052144 (rank : 53) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8ND99 | Gene names | C19orf16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C19orf16. | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.051769 (rank : 54) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.051701 (rank : 55) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.051645 (rank : 56) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.051038 (rank : 57) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.050963 (rank : 58) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.050848 (rank : 59) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.050485 (rank : 60) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
RIN3_HUMAN
|
||||||
| NC score | 0.046138 (rank : 61) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8TB24, Q8NF30, Q8TEE8, Q8WYP4, Q9H6A5, Q9HAG1 | Gene names | RIN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras and Rab interactor 3 (Ras interaction/interference protein 3). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.044746 (rank : 62) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.044556 (rank : 63) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.044362 (rank : 64) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.043723 (rank : 65) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.043002 (rank : 66) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.042825 (rank : 67) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
RBM4_HUMAN
|
||||||
| NC score | 0.042542 (rank : 68) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWF3, O02916, Q6P1P2, Q8WU85 | Gene names | RBM4, RBM4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (hLark). | |||||
|
RBM4_MOUSE
|
||||||
| NC score | 0.042476 (rank : 69) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C7Q4, O08752, Q8BN66 | Gene names | Rbm4, Rbm4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4 (RNA-binding motif protein 4) (RNA-binding motif protein 4a) (Lark homolog) (mLark). | |||||
|
RBM4B_HUMAN
|
||||||
| NC score | 0.042331 (rank : 70) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BQ04 | Gene names | RBM4B, RBM30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
RBM4B_MOUSE
|
||||||
| NC score | 0.042275 (rank : 71) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VE92 | Gene names | Rbm4b, Rbm30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 4B (RNA-binding motif protein 4B) (RNA-binding protein 30) (RNA-binding motif protein 30). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.041718 (rank : 72) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.041317 (rank : 73) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.039718 (rank : 74) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.039672 (rank : 75) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.039474 (rank : 76) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.038948 (rank : 77) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.036570 (rank : 78) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
COIA1_HUMAN
|
||||||
| NC score | 0.035902 (rank : 79) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39060, Q9UK38, Q9Y6Q7, Q9Y6Q8 | Gene names | COL18A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
OGFR_MOUSE
|
||||||
| NC score | 0.033190 (rank : 80) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99PG2, Q99L33 | Gene names | Ogfr | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.031763 (rank : 81) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.031085 (rank : 82) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
RECQ4_MOUSE
|
||||||
| NC score | 0.028898 (rank : 83) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q75NR7, Q76MT1, Q99PV9 | Gene names | Recql4, Recq4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent DNA helicase Q4 (EC 3.6.1.-) (RecQ protein-like 4). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.028672 (rank : 84) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.028286 (rank : 85) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
CPSF4_MOUSE
|
||||||
| NC score | 0.028105 (rank : 86) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BQZ5, O54930 | Gene names | Cpsf4, Cpsf30 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF 30 kDa subunit) (Clipper homolog) (Clipper/CPSF 30K). | |||||
|
CSPG5_MOUSE
|
||||||
| NC score | 0.026870 (rank : 87) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q71M36, Q71M37, Q7TNT8, Q8BPJ5, Q9QY32 | Gene names | Cspg5, Caleb, Ngc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
SOX5_HUMAN
|
||||||
| NC score | 0.026426 (rank : 88) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35711, Q86UK8, Q8J017, Q8J018, Q8J019, Q8J020, Q8N1D9, Q8N7E0, Q8TEA4 | Gene names | SOX5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
MYO15_HUMAN
|
||||||
| NC score | 0.026422 (rank : 89) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UKN7 | Gene names | MYO15A, MYO15 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-15 (Myosin XV) (Unconventional myosin-15). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.026251 (rank : 90) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SOX5_MOUSE
|
||||||
| NC score | 0.026233 (rank : 91) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35710, O88184, O89018 | Gene names | Sox5, Sox-5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-5. | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.025689 (rank : 92) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.025377 (rank : 93) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
ZN395_HUMAN
|
||||||
| NC score | 0.024928 (rank : 94) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8N7, Q6F6H2, Q9BY72, Q9NPB2, Q9NS57, Q9NS58, Q9NS59 | Gene names | ZNF395, HDBP2, PBF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 395 (Papillomavirus-binding factor) (Papillomavirus regulatory factor 1) (PRF-1) (Huntington disease gene regulatory region-binding protein 2) (HDBP-2) (HD gene regulatory region-binding protein 2) (HD-regulating factor 2) (HDRF-2). | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.024760 (rank : 95) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
LMBL3_MOUSE
|
||||||
| NC score | 0.024714 (rank : 96) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BLB7, Q641L7, Q6ZPI2, Q8C0G4 | Gene names | L3mbtl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(3)malignant brain tumor-like 3 protein (L(3)mbt-like 3 protein). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.024537 (rank : 97) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.024073 (rank : 98) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.023312 (rank : 99) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.023181 (rank : 100) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
ZN409_HUMAN
|
||||||
| NC score | 0.022941 (rank : 101) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UPU6 | Gene names | ZNF409, KIAA1056 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 409. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.022747 (rank : 102) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
AOC2_HUMAN
|
||||||
| NC score | 0.021825 (rank : 103) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75106, O00120, O75105, Q4TTW5, Q9UNY0 | Gene names | AOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retina-specific copper amine oxidase precursor (EC 1.4.3.6) (RAO) (Amine oxidase [copper-containing]). | |||||
|
AOC3_HUMAN
|
||||||
| NC score | 0.021825 (rank : 104) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16853, Q45F94 | Gene names | AOC3, VAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane copper amine oxidase (EC 1.4.3.6) (Semicarbazide-sensitive amine oxidase) (SSAO) (Vascular adhesion protein 1) (VAP-1) (HPAO). | |||||
|
NUMBL_MOUSE
|
||||||
| NC score | 0.020643 (rank : 105) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O08919, Q6NVG8 | Gene names | Numbl, Nbl | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein. | |||||
|
FAIM2_HUMAN
|
||||||
| NC score | 0.020167 (rank : 106) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BWQ8, Q9UJY9, Q9Y2F7 | Gene names | FAIM2, KIAA0950, LFG, TMBIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Fas apoptotic inhibitory molecule 2 (Lifeguard protein) (Transmembrane BAX inhibitor motif-containing protein 2). | |||||
|
RASF7_MOUSE
|
||||||
| NC score | 0.019703 (rank : 107) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DD19 | Gene names | Rassf7 | |||
|
Domain Architecture |

|
|||||
| Description | Ras association domain-containing protein 7. | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.019365 (rank : 108) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
SCAP_HUMAN
|
||||||
| NC score | 0.019251 (rank : 109) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q12770, Q8N2E0, Q8WUA1 | Gene names | SCAP, KIAA0199 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein cleavage-activating protein (SREBP cleavage-activating protein) (SCAP). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.019101 (rank : 110) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.019072 (rank : 111) | θ value | 0.0148317 (rank : 5) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ANR11_HUMAN
|
||||||
| NC score | 0.018246 (rank : 112) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UB99, Q6NTG1, Q6QMF8 | Gene names | ANKRD11, ANCO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 11 (Ankyrin repeat-containing cofactor 1). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.018141 (rank : 113) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.018122 (rank : 114) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
BI2L1_HUMAN
|
||||||
| NC score | 0.017997 (rank : 115) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UHR4, Q75L21, Q75L22, Q96CV4, Q9Y2M8 | Gene names | BAIAP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAI1-associated protein 2-like protein 1). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.017385 (rank : 116) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
ERF_MOUSE
|
||||||
| NC score | 0.016992 (rank : 117) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70459 | Gene names | Erf | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF. | |||||
|
HXD1_MOUSE
|
||||||
| NC score | 0.016473 (rank : 118) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01822 | Gene names | Hoxd1, Hox-4.9, Hoxd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1 (Hox-4.9). | |||||
|
ERF_HUMAN
|
||||||
| NC score | 0.015698 (rank : 119) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P50548, Q9UPI7 | Gene names | ERF | |||
|
Domain Architecture |

|
|||||
| Description | ETS domain-containing transcription factor ERF (Ets2 repressor factor). | |||||
|
HXD1_HUMAN
|
||||||
| NC score | 0.015643 (rank : 120) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9GZZ0 | Gene names | HOXD1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D1. | |||||
|
SPAG1_HUMAN
|
||||||
| NC score | 0.015326 (rank : 121) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07617, Q7Z5G1 | Gene names | SPAG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 1 (Infertility-related sperm protein Spag-1) (HSD-3.8). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.015298 (rank : 122) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.014964 (rank : 123) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.013953 (rank : 124) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.013716 (rank : 125) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.013165 (rank : 126) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.010515 (rank : 127) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
HXB13_HUMAN
|
||||||
| NC score | 0.009992 (rank : 128) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92826, Q96QM4, Q99810 | Gene names | HOXB13 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B13. | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.009723 (rank : 129) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
ZSCA5_HUMAN
|
||||||
| NC score | 0.008883 (rank : 130) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
PAX9_MOUSE
|
||||||
| NC score | 0.008254 (rank : 131) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P47242 | Gene names | Pax9, Pax-9 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-9. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.007727 (rank : 132) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
NEO1_MOUSE
|
||||||
| NC score | 0.007464 (rank : 133) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
MOES_MOUSE
|
||||||
| NC score | 0.004899 (rank : 134) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 923 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P26041, Q3UL28, Q8BSN4 | Gene names | Msn | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
ZN688_HUMAN
|
||||||
| NC score | 0.004865 (rank : 135) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 873 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WV14, O75701, Q8IW91, Q96MN0 | Gene names | ZNF688 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 688. | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.003451 (rank : 136) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
ZN384_HUMAN
|
||||||
| NC score | 0.003358 (rank : 137) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TF68, O15407, Q8N938 | Gene names | ZNF384, CAGH1, CIZ, NMP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 384 (Nuclear matrix transcription factor 4) (Nuclear matrix protein 4) (CAG repeat protein 1) (CAS-interacting zinc finger protein). | |||||
|
PMYT1_MOUSE
|
||||||
| NC score | 0.003184 (rank : 138) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 95 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ESG9, Q8R3L4 | Gene names | Pkmyt1, Myt1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase (EC 2.7.11.1) (Myt1 kinase). | |||||