Please be patient as the page loads
|
RNF37_HUMAN
|
||||||
| SwissProt Accessions | O94941 | Gene names | UBOX5, KIAA0860, RNF37, UBCE7IP5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (U-box domain-containing protein 5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RNF37_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O94941 | Gene names | UBOX5, KIAA0860, RNF37, UBCE7IP5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
RNF37_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.965856 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q925F4 | Gene names | Ubox5, Rnf37, Ubce7ip5, Uip5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (UbcM4-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
UBE4A_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.228756 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14139, Q6P5T4, Q7Z639 | Gene names | UBE4A, KIAA0126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin conjugation factor E4 A. | |||||
|
UBE4B_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 4) | NC score | 0.198902 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
UBE4B_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 5) | NC score | 0.206276 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
TBCE_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.068251 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15813 | Gene names | TBCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone E (Tubulin-folding cofactor E). | |||||
|
RN168_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.155309 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
RN168_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 8) | NC score | 0.150603 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80XJ2 | Gene names | Rnf168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.035686 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NHLC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.137451 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BR37 | Gene names | Nhlrc1, Epm2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NHL repeat-containing protein 1 (Malin). | |||||
|
TRI65_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.099400 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BFW4, Q6PGD3 | Gene names | Trim65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 65. | |||||
|
CBLC_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.136450 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULV8, Q8N1E5, Q9Y5Z2, Q9Y5Z3 | Gene names | CBLC, CBL3, RNF57 | |||
|
Domain Architecture |
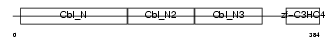
|
|||||
| Description | Signal transduction protein CBL-C (SH3-binding protein CBL-C) (CBL-3) (RING finger protein 57). | |||||
|
ZN313_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.138299 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ET26, Q3UFU8, Q8K5A2 | Gene names | Znf313, Zfp228, Zfp313, Znf228 | |||
|
Domain Architecture |
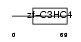
|
|||||
| Description | Zinc finger protein 313. | |||||
|
CBLC_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.130540 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80XL1, Q99PB6, Q9D6L2, Q9D7A9 | Gene names | Cblc, Cbl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transduction protein CBL-C (SH3-binding protein CBL-C) (CBL-3). | |||||
|
PRD14_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.012233 (rank : 97) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9GZV8, Q86UX9 | Gene names | PRDM14 | |||
|
Domain Architecture |
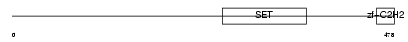
|
|||||
| Description | PR domain zinc finger protein 14 (PR domain-containing protein 14). | |||||
|
TRI43_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.070564 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96BQ3 | Gene names | TRIM43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 43. | |||||
|
ZN364_HUMAN
|
||||||
| θ value | 0.365318 (rank : 17) | NC score | 0.092198 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y4L5, Q5T2V9, Q7Z2J2 | Gene names | ZNF364, RNF115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 364 (Rabring 7) (RING finger protein 115). | |||||
|
ATF5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.057863 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
RNF17_MOUSE
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.096981 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MV7 | Gene names | Rnf17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
ZN313_HUMAN
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.126546 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y508, Q6N0B0 | Gene names | ZNF313, RNF114, ZNF228 | |||
|
Domain Architecture |
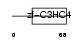
|
|||||
| Description | Zinc finger protein 313 (RING finger protein 114). | |||||
|
MGR4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.017163 (rank : 90) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
RN125_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.118237 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96EQ8, Q9NX39 | Gene names | RNF125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-) (T-cell RING activation protein 1) (TRAC-1). | |||||
|
TRI68_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.050861 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K243 | Gene names | Trim68, Rnf137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif containing protein 68 (Ring finger protein 137). | |||||
|
ZN364_MOUSE
|
||||||
| θ value | 0.62314 (rank : 24) | NC score | 0.089729 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D0C1, Q8R5A1, Q9D885 | Gene names | Znf364, Zfp364 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 364 (Rabring 7). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.009333 (rank : 102) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
MTF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 26) | NC score | 0.009309 (rank : 103) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07243 | Gene names | Mtf1, Mtf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.017016 (rank : 91) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
STUB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.079452 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
STUB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.080885 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
TRAF2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.072839 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12933, Q96NT2 | Gene names | TRAF2, TRAP3 | |||
|
Domain Architecture |
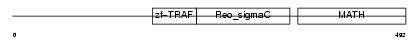
|
|||||
| Description | TNF receptor-associated factor 2 (Tumor necrosis factor type 2 receptor-associated protein 3). | |||||
|
TRAF2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.074518 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P39429, O54896 | Gene names | Traf2 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2. | |||||
|
TRAF3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.073535 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13114, Q12990, Q13076, Q13947, Q6AZX1, Q9UNL1 | Gene names | TRAF3, CAP1, CRAF1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (CD40-binding protein) (CD40BP) (LMP1-associated protein) (LAP1) (CAP-1). | |||||
|
TRIM7_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.050852 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
RN122_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.052412 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H9V4, Q52LK3 | Gene names | RNF122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
TRAF3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.072691 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60803, Q62380 | Gene names | Traf3, Craf1, Trafamn | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (TRAFAMN). | |||||
|
TNR8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.032736 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
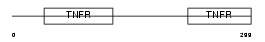
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
CNTRB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.032791 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.047942 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI50_MOUSE
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.059600 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q810I2 | Gene names | Trim50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 50. | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.071135 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.070760 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
CTR2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.032167 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.050836 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
TRI25_MOUSE
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.084595 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61510 | Gene names | Trim25, Efp, Zfp147, Znf147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp). | |||||
|
CBL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.102640 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CBL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.102530 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
NHLC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.090674 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6VVB1, Q6IMH1 | Gene names | NHLRC1, EPM2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NHL repeat-containing protein 1 (Malin). | |||||
|
RN122_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.048465 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP31, Q80VA7, Q8BGD3 | Gene names | Rnf122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
RN185_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.096906 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YT2, Q6ZWS3 | Gene names | Rnf185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.011040 (rank : 100) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
TRI25_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.080357 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
TRI42_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.054007 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
TRI47_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.072695 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
RNF5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.091568 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.013035 (rank : 95) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
TRI42_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.052411 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
CBLB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.097228 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13191, Q13192, Q13193, Q3LIC0, Q8IVC5 | Gene names | CBLB, RNF56 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b) (RING finger protein 56). | |||||
|
CBLB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.097275 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
EMBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.019122 (rank : 89) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |
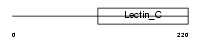
|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
PIWL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.011798 (rank : 98) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDG1, Q3TQE8, Q99MV6, Q9JMB6 | Gene names | Piwil2, Mili | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
RBM16_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.022937 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
SN1L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.000416 (rank : 107) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60670 | Gene names | Snf1lk, Msk | |||
|
Domain Architecture |
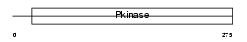
|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK) (HRT-20) (Myocardial SNF1- like kinase). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.044607 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.001855 (rank : 106) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ARD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.019565 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 621 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P36406, Q9BZY4, Q9BZY5 | Gene names | TRIM23, ARD1, ARFD1, RNF46 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23) (RING finger protein 46). | |||||
|
ARD1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.020840 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BGX0, Q8C2B6, Q8CDA4, Q8CDA7 | Gene names | Trim23, Arfd1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23). | |||||
|
ARHGF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.011313 (rank : 99) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.012979 (rank : 96) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
PRGC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.014802 (rank : 94) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
RN185_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.100637 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96GF1, Q8N900 | Gene names | RNF185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
RNF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.088508 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.009595 (rank : 101) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SPON2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.016668 (rank : 92) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUD6, Q9ULW1 | Gene names | SPON2, DIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-2 precursor (Mindin) (Differentially expressed in cancerous and non-cancerous lung cells 1) (DIL-1). | |||||
|
TRI54_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.048754 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYV2, Q53SY4, Q9BYV3 | Gene names | TRIM54, MURF, MURF3, RNF30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 54 (Muscle-specific RING finger protein 3) (MuRF3) (MURF-3) (Muscle-specific RING finger protein) (MuRF) (RING finger protein 30). | |||||
|
TRI54_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.048783 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERP3 | Gene names | Trim54, Murf, Rnf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 54 (Muscle-specific RING finger protein 3) (MuRF3) (MURF-3) (Muscle-specific RING finger protein) (MuRF) (RING finger protein 30). | |||||
|
TRI65_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.077985 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PJ69, Q4G0F0, Q6DKJ6, Q9BRP6 | Gene names | TRIM65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 65. | |||||
|
ZF276_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.005611 (rank : 105) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
ATX1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.016145 (rank : 93) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.005951 (rank : 104) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | -0.000016 (rank : 108) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TRI68_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.042837 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6AZZ1, Q8WZ70, Q96PF7, Q9H9C2, Q9NW18 | Gene names | TRIM68, RNF137, SS56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 68 (RING finger protein 137) (SSA protein SS-56) (SS-56). | |||||
|
TRIM7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.046957 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q923T7, Q5NCB6, Q99PQ5 | Gene names | Trim7, Gnip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein). | |||||
|
AMFR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.052667 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKV5, Q8IZ70 | Gene names | AMFR | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor, isoform 2 (EC 6.3.2.-) (AMF receptor) (gp78). | |||||
|
AMFR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051069 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R049, Q8K008, Q99LH5 | Gene names | Amfr | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor (EC 6.3.2.19) (AMF receptor). | |||||
|
BFAR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052100 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
BFAR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.063195 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
PEX10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.066752 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |
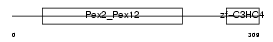
|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
RN125_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.085863 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
RN126_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.064776 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RN126_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.072011 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
RN146_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.051336 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NTX7, Q6FIB2, Q7L8H4, Q96K03, Q9NTX6 | Gene names | RNF146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 146 (Dactylidin). | |||||
|
RN146_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050959 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CZW6, Q3TF93 | Gene names | Rnf146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 146. | |||||
|
RN166_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.075445 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96A37, Q96DM0 | Gene names | RNF166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
RN166_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.075575 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q3U9F6 | Gene names | Rnf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
RN169_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.062926 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
RNF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.051442 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QZS2, O35941, Q541Z6 | Gene names | Rnf4 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 4. | |||||
|
TRAF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.060757 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y4K3, Q8NEH5 | Gene names | TRAF6, RNF85 | |||
|
Domain Architecture |
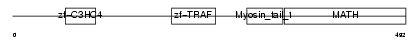
|
|||||
| Description | TNF receptor-associated factor 6 (Interleukin 1 signal transducer) (RING finger protein 85). | |||||
|
TRAF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.060264 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70196, Q8BLV2 | Gene names | Traf6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 6. | |||||
|
TRI32_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.053372 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
TRI32_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.052952 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
TRI40_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.059376 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P9F5, Q8TD96 | Gene names | TRIM40, RNF35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 40 (RING finger protein 35). | |||||
|
TRI47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.063241 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
TRI50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.050974 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86XT4, Q86XT3 | Gene names | TRIM50, TRIM50A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 50. | |||||
|
TRI56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.059904 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
TRI56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.058302 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80VI1, Q8CAY0 | Gene names | Trim56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56. | |||||
|
TRI73_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050804 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UV7, Q8N0S3 | Gene names | TRIM73, TRIM50B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 73 (Tripartite motif-containing protein 50B). | |||||
|
TRIM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.051541 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
TRIM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051058 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
RNF37_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | O94941 | Gene names | UBOX5, KIAA0860, RNF37, UBCE7IP5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
RNF37_MOUSE
|
||||||
| NC score | 0.965856 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q925F4 | Gene names | Ubox5, Rnf37, Ubce7ip5, Uip5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (UbcM4-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
UBE4A_HUMAN
|
||||||
| NC score | 0.228756 (rank : 3) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14139, Q6P5T4, Q7Z639 | Gene names | UBE4A, KIAA0126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin conjugation factor E4 A. | |||||
|
UBE4B_MOUSE
|
||||||
| NC score | 0.206276 (rank : 4) | θ value | 0.00665767 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ES00, Q9EQE0 | Gene names | Ube4b, Ufd2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Ufd2a). | |||||
|
UBE4B_HUMAN
|
||||||
| NC score | 0.198902 (rank : 5) | θ value | 0.00665767 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95155, O75169, O95338, Q5SZ12, Q5SZ16, Q96QD4, Q9BYI7 | Gene names | UBE4B, HDNB1, KIAA0684, UFD2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin conjugation factor E4 B (Ubiquitin fusion degradation protein 2) (Homozygously deleted in neuroblastoma 1). | |||||
|
RN168_HUMAN
|
||||||
| NC score | 0.155309 (rank : 6) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
RN168_MOUSE
|
||||||
| NC score | 0.150603 (rank : 7) | θ value | 0.0431538 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q80XJ2 | Gene names | Rnf168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
ZN313_MOUSE
|
||||||
| NC score | 0.138299 (rank : 8) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9ET26, Q3UFU8, Q8K5A2 | Gene names | Znf313, Zfp228, Zfp313, Znf228 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 313. | |||||
|
NHLC1_MOUSE
|
||||||
| NC score | 0.137451 (rank : 9) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BR37 | Gene names | Nhlrc1, Epm2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NHL repeat-containing protein 1 (Malin). | |||||
|
CBLC_HUMAN
|
||||||
| NC score | 0.136450 (rank : 10) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9ULV8, Q8N1E5, Q9Y5Z2, Q9Y5Z3 | Gene names | CBLC, CBL3, RNF57 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transduction protein CBL-C (SH3-binding protein CBL-C) (CBL-3) (RING finger protein 57). | |||||
|
CBLC_MOUSE
|
||||||
| NC score | 0.130540 (rank : 11) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80XL1, Q99PB6, Q9D6L2, Q9D7A9 | Gene names | Cblc, Cbl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transduction protein CBL-C (SH3-binding protein CBL-C) (CBL-3). | |||||
|
ZN313_HUMAN
|
||||||
| NC score | 0.126546 (rank : 12) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9Y508, Q6N0B0 | Gene names | ZNF313, RNF114, ZNF228 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 313 (RING finger protein 114). | |||||
|
RN125_HUMAN
|
||||||
| NC score | 0.118237 (rank : 13) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96EQ8, Q9NX39 | Gene names | RNF125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-) (T-cell RING activation protein 1) (TRAC-1). | |||||
|
CBL_HUMAN
|
||||||
| NC score | 0.102640 (rank : 14) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P22681 | Gene names | CBL, CBL2, RNF55 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene) (RING finger protein 55). | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.102530 (rank : 15) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
RN185_HUMAN
|
||||||
| NC score | 0.100637 (rank : 16) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96GF1, Q8N900 | Gene names | RNF185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
TRI65_MOUSE
|
||||||
| NC score | 0.099400 (rank : 17) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BFW4, Q6PGD3 | Gene names | Trim65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 65. | |||||
|
CBLB_MOUSE
|
||||||
| NC score | 0.097275 (rank : 18) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q3TTA7 | Gene names | Cblb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b). | |||||
|
CBLB_HUMAN
|
||||||
| NC score | 0.097228 (rank : 19) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13191, Q13192, Q13193, Q3LIC0, Q8IVC5 | Gene names | CBLB, RNF56 | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL-B (EC 6.3.2.-) (Signal transduction protein CBL-B) (SH3-binding protein CBL-B) (Casitas B-lineage lymphoma proto-oncogene b) (RING finger protein 56). | |||||
|
RNF17_MOUSE
|
||||||
| NC score | 0.096981 (rank : 20) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MV7 | Gene names | Rnf17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
RN185_MOUSE
|
||||||
| NC score | 0.096906 (rank : 21) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q91YT2, Q6ZWS3 | Gene names | Rnf185 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 185. | |||||
|
ZN364_HUMAN
|
||||||
| NC score | 0.092198 (rank : 22) | θ value | 0.365318 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y4L5, Q5T2V9, Q7Z2J2 | Gene names | ZNF364, RNF115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 364 (Rabring 7) (RING finger protein 115). | |||||
|
RNF5_MOUSE
|
||||||
| NC score | 0.091568 (rank : 23) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
NHLC1_HUMAN
|
||||||
| NC score | 0.090674 (rank : 24) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6VVB1, Q6IMH1 | Gene names | NHLRC1, EPM2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NHL repeat-containing protein 1 (Malin). | |||||
|
ZN364_MOUSE
|
||||||
| NC score | 0.089729 (rank : 25) | θ value | 0.62314 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D0C1, Q8R5A1, Q9D885 | Gene names | Znf364, Zfp364 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 364 (Rabring 7). | |||||
|
RNF5_HUMAN
|
||||||
| NC score | 0.088508 (rank : 26) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
RN125_MOUSE
|
||||||
| NC score | 0.085863 (rank : 27) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
TRI25_MOUSE
|
||||||
| NC score | 0.084595 (rank : 28) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61510 | Gene names | Trim25, Efp, Zfp147, Znf147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp). | |||||
|
STUB1_MOUSE
|
||||||
| NC score | 0.080885 (rank : 29) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WUD1, Q9DCJ0 | Gene names | Stub1, Chip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP). | |||||
|
TRI25_HUMAN
|
||||||
| NC score | 0.080357 (rank : 30) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
STUB1_HUMAN
|
||||||
| NC score | 0.079452 (rank : 31) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UNE7, O60526, Q969U2, Q9HBT1 | Gene names | STUB1, CHIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STIP1 homology and U box-containing protein 1 (EC 6.3.2.-) (STIP1 homology and U-box-containing protein 1) (Carboxy terminus of Hsp70- interacting protein) (E3 ubiquitin protein ligase CHIP) (CLL- associated antigen KW-8) (Antigen NY-CO-7). | |||||
|
TRI65_HUMAN
|
||||||
| NC score | 0.077985 (rank : 32) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q6PJ69, Q4G0F0, Q6DKJ6, Q9BRP6 | Gene names | TRIM65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 65. | |||||
|
RN166_MOUSE
|
||||||
| NC score | 0.075575 (rank : 33) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q3U9F6 | Gene names | Rnf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
RN166_HUMAN
|
||||||
| NC score | 0.075445 (rank : 34) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96A37, Q96DM0 | Gene names | RNF166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
TRAF2_MOUSE
|
||||||
| NC score | 0.074518 (rank : 35) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P39429, O54896 | Gene names | Traf2 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2. | |||||
|
TRAF3_HUMAN
|
||||||
| NC score | 0.073535 (rank : 36) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13114, Q12990, Q13076, Q13947, Q6AZX1, Q9UNL1 | Gene names | TRAF3, CAP1, CRAF1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (CD40-binding protein) (CD40BP) (LMP1-associated protein) (LAP1) (CAP-1). | |||||
|
TRAF2_HUMAN
|
||||||
| NC score | 0.072839 (rank : 37) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q12933, Q96NT2 | Gene names | TRAF2, TRAP3 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2 (Tumor necrosis factor type 2 receptor-associated protein 3). | |||||
|
TRI47_MOUSE
|
||||||
| NC score | 0.072695 (rank : 38) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
TRAF3_MOUSE
|
||||||
| NC score | 0.072691 (rank : 39) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q60803, Q62380 | Gene names | Traf3, Craf1, Trafamn | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (TRAFAMN). | |||||
|
RN126_MOUSE
|
||||||
| NC score | 0.072011 (rank : 40) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.071135 (rank : 41) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.070760 (rank : 42) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRI43_HUMAN
|
||||||
| NC score | 0.070564 (rank : 43) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96BQ3 | Gene names | TRIM43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 43. | |||||
|
TBCE_HUMAN
|
||||||
| NC score | 0.068251 (rank : 44) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15813 | Gene names | TBCE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin-specific chaperone E (Tubulin-folding cofactor E). | |||||
|
PEX10_HUMAN
|
||||||
| NC score | 0.066752 (rank : 45) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
RN126_HUMAN
|
||||||
| NC score | 0.064776 (rank : 46) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
TRI47_HUMAN
|
||||||
| NC score | 0.063241 (rank : 47) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
BFAR_MOUSE
|
||||||
| NC score | 0.063195 (rank : 48) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
RN169_HUMAN
|
||||||
| NC score | 0.062926 (rank : 49) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NCN4, Q6N015 | Gene names | RNF169, KIAA1991 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 169. | |||||
|
TRAF6_HUMAN
|
||||||
| NC score | 0.060757 (rank : 50) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y4K3, Q8NEH5 | Gene names | TRAF6, RNF85 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 6 (Interleukin 1 signal transducer) (RING finger protein 85). | |||||
|
TRAF6_MOUSE
|
||||||
| NC score | 0.060264 (rank : 51) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70196, Q8BLV2 | Gene names | Traf6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 6. | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.059904 (rank : 52) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
TRI50_MOUSE
|
||||||
| NC score | 0.059600 (rank : 53) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q810I2 | Gene names | Trim50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 50. | |||||
|
TRI40_HUMAN
|
||||||
| NC score | 0.059376 (rank : 54) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P9F5, Q8TD96 | Gene names | TRIM40, RNF35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 40 (RING finger protein 35). | |||||
|
TRI56_MOUSE
|
||||||
| NC score | 0.058302 (rank : 55) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80VI1, Q8CAY0 | Gene names | Trim56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56. | |||||
|
ATF5_MOUSE
|
||||||
| NC score | 0.057863 (rank : 56) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70191, Q58E51, Q99NH6 | Gene names | Atf5, Atfx, Nap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5-alpha/beta) (Transcription factor ATFx) (Transcription factor-like protein ODA-10) (BZIP protein ATF7) (NRIF3- associated protein) (NAP1). | |||||
|
TRI42_HUMAN
|
||||||
| NC score | 0.054007 (rank : 57) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
TRI32_HUMAN
|
||||||
| NC score | 0.053372 (rank : 58) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
TRI32_MOUSE
|
||||||
| NC score | 0.052952 (rank : 59) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
AMFR2_HUMAN
|
||||||
| NC score | 0.052667 (rank : 60) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UKV5, Q8IZ70 | Gene names | AMFR | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor, isoform 2 (EC 6.3.2.-) (AMF receptor) (gp78). | |||||
|
RN122_HUMAN
|
||||||
| NC score | 0.052412 (rank : 61) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H9V4, Q52LK3 | Gene names | RNF122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
TRI42_MOUSE
|
||||||
| NC score | 0.052411 (rank : 62) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9D2H5 | Gene names | Trim42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
BFAR_HUMAN
|
||||||
| NC score | 0.052100 (rank : 63) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
TRIM2_HUMAN
|
||||||
| NC score | 0.051541 (rank : 64) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
RNF4_MOUSE
|
||||||
| NC score | 0.051442 (rank : 65) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QZS2, O35941, Q541Z6 | Gene names | Rnf4 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 4. | |||||
|
RN146_HUMAN
|
||||||
| NC score | 0.051336 (rank : 66) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NTX7, Q6FIB2, Q7L8H4, Q96K03, Q9NTX6 | Gene names | RNF146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 146 (Dactylidin). | |||||
|
AMFR_MOUSE
|
||||||
| NC score | 0.051069 (rank : 67) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9R049, Q8K008, Q99LH5 | Gene names | Amfr | |||
|
Domain Architecture |

|
|||||
| Description | Autocrine motility factor receptor (EC 6.3.2.19) (AMF receptor). | |||||
|
TRIM2_MOUSE
|
||||||
| NC score | 0.051058 (rank : 68) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
TRI50_HUMAN
|
||||||
| NC score | 0.050974 (rank : 69) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q86XT4, Q86XT3 | Gene names | TRIM50, TRIM50A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 50. | |||||
|
RN146_MOUSE
|
||||||
| NC score | 0.050959 (rank : 70) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CZW6, Q3TF93 | Gene names | Rnf146 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 146. | |||||
|
TRI68_MOUSE
|
||||||
| NC score | 0.050861 (rank : 71) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K243 | Gene names | Trim68, Rnf137 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif containing protein 68 (Ring finger protein 137). | |||||
|
TRIM7_HUMAN
|
||||||
| NC score | 0.050852 (rank : 72) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C029, Q969F5, Q96F67, Q96J89, Q96J90 | Gene names | TRIM7, GNIP, RNF90 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein) (RING finger protein 90). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.050836 (rank : 73) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
TRI73_HUMAN
|
||||||
| NC score | 0.050804 (rank : 74) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86UV7, Q8N0S3 | Gene names | TRIM73, TRIM50B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 73 (Tripartite motif-containing protein 50B). | |||||
|
TRI54_MOUSE
|
||||||
| NC score | 0.048783 (rank : 75) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ERP3 | Gene names | Trim54, Murf, Rnf30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 54 (Muscle-specific RING finger protein 3) (MuRF3) (MURF-3) (Muscle-specific RING finger protein) (MuRF) (RING finger protein 30). | |||||
|
TRI54_HUMAN
|
||||||
| NC score | 0.048754 (rank : 76) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9BYV2, Q53SY4, Q9BYV3 | Gene names | TRIM54, MURF, MURF3, RNF30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 54 (Muscle-specific RING finger protein 3) (MuRF3) (MURF-3) (Muscle-specific RING finger protein) (MuRF) (RING finger protein 30). | |||||
|
RN122_MOUSE
|
||||||
| NC score | 0.048465 (rank : 77) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BP31, Q80VA7, Q8BGD3 | Gene names | Rnf122 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 122. | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.047942 (rank : 78) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRIM7_MOUSE
|
||||||
| NC score | 0.046957 (rank : 79) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q923T7, Q5NCB6, Q99PQ5 | Gene names | Trim7, Gnip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 7 (Glycogenin-interacting protein). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.044607 (rank : 80) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TRI68_HUMAN
|
||||||
| NC score | 0.042837 (rank : 81) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6AZZ1, Q8WZ70, Q96PF7, Q9H9C2, Q9NW18 | Gene names | TRIM68, RNF137, SS56 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 68 (RING finger protein 137) (SSA protein SS-56) (SS-56). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.035686 (rank : 82) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
CNTRB_HUMAN
|
||||||
| NC score | 0.032791 (rank : 83) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N137, Q331K3, Q69YV7, Q8NCB8, Q8WXV3, Q96CQ7, Q9C060 | Gene names | CNTROB, LIP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.032736 (rank : 84) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
CTR2_MOUSE
|
||||||
| NC score | 0.032167 (rank : 85) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18581 | Gene names | Slc7a2, Atrc2, Tea | |||
|
Domain Architecture |

|
|||||
| Description | Low-affinity cationic amino acid transporter 2 (CAT-2) (CAT2) (TEA protein) (T-cell early activation protein) (20.5). | |||||
|
RBM16_HUMAN
|
||||||
| NC score | 0.022937 (rank : 86) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPN6, Q5TBU6, Q9BQN8, Q9BX43 | Gene names | RBM16, KIAA1116 | |||
|
Domain Architecture |

|
|||||
| Description | Putative RNA-binding protein 16 (RNA-binding motif protein 16). | |||||
|
ARD1_MOUSE
|
||||||
| NC score | 0.020840 (rank : 87) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8BGX0, Q8C2B6, Q8CDA4, Q8CDA7 | Gene names | Trim23, Arfd1 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23). | |||||
|
ARD1_HUMAN
|
||||||
| NC score | 0.019565 (rank : 88) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 621 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P36406, Q9BZY4, Q9BZY5 | Gene names | TRIM23, ARD1, ARFD1, RNF46 | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein ARD-1 (ADP-ribosylation factor domain protein 1) (Tripartite motif-containing protein 23) (RING finger protein 46). | |||||
|
EMBP_MOUSE
|
||||||
| NC score | 0.019122 (rank : 89) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61878 | Gene names | Prg2, Mbp-1 | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil granule major basic protein precursor (MBP) (Proteoglycan 2, bone marrow). | |||||
|
MGR4_HUMAN
|
||||||
| NC score | 0.017163 (rank : 90) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14833, Q5SZ84 | Gene names | GRM4, GPRC1D, MGLUR4 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 4 precursor (mGluR4). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.017016 (rank : 91) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
SPON2_HUMAN
|
||||||
| NC score | 0.016668 (rank : 92) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUD6, Q9ULW1 | Gene names | SPON2, DIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-2 precursor (Mindin) (Differentially expressed in cancerous and non-cancerous lung cells 1) (DIL-1). | |||||
|
ATX1_MOUSE
|
||||||
| NC score | 0.016145 (rank : 93) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P54254 | Gene names | Atxn1, Sca1 | |||
|
Domain Architecture |

|
|||||
| Description | Ataxin-1 (Spinocerebellar ataxia type 1 protein homolog). | |||||
|
PRGC2_HUMAN
|
||||||
| NC score | 0.014802 (rank : 94) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86YN6, Q86YN3, Q86YN4, Q86YN5, Q8N1N9, Q8TDE4, Q8TDE5 | Gene names | PPARGC1B, PERC, PGC1, PGC1B, PPARGC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPAR gamma coactivator-1beta) (PGC-1-related estrogen receptor alpha coactivator) (PPARGC-1-beta) (PGC-1-beta). | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.013035 (rank : 95) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.012979 (rank : 96) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
PRD14_HUMAN
|
||||||
| NC score | 0.012233 (rank : 97) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9GZV8, Q86UX9 | Gene names | PRDM14 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 14 (PR domain-containing protein 14). | |||||
|
PIWL2_MOUSE
|
||||||
| NC score | 0.011798 (rank : 98) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CDG1, Q3TQE8, Q99MV6, Q9JMB6 | Gene names | Piwil2, Mili | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Piwi-like protein 2. | |||||
|
ARHGF_HUMAN
|
||||||
| NC score | 0.011313 (rank : 99) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94989, Q8N449, Q9H8B4 | Gene names | ARHGEF15, KIAA0915 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 15 (Vsm-RhoGEF). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.011040 (rank : 100) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.009595 (rank : 101) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | 0.009333 (rank : 102) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
MTF1_MOUSE
|
||||||
| NC score | 0.009309 (rank : 103) | θ value | 0.813845 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07243 | Gene names | Mtf1, Mtf-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.005951 (rank : 104) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
ZF276_HUMAN
|
||||||
| NC score | 0.005611 (rank : 105) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.001855 (rank : 106) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SN1L1_MOUSE
|
||||||
| NC score | 0.000416 (rank : 107) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60670 | Gene names | Snf1lk, Msk | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase SNF1-like kinase 1 (EC 2.7.11.1) (Serine/threonine-protein kinase SNF1LK) (HRT-20) (Myocardial SNF1- like kinase). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.000016 (rank : 108) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||