Please be patient as the page loads
|
FURIN_MOUSE
|
||||||
| SwissProt Accessions | P23188 | Gene names | Furin, Fur, Pcsk3 | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme) (Prohormone convertase 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FURIN_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998706 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P09958, Q14336, Q6LBS3 | Gene names | FURIN, FUR, PACE | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme). | |||||
|
FURIN_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P23188 | Gene names | Furin, Fur, Pcsk3 | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme) (Prohormone convertase 3). | |||||
|
PCSK4_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.990565 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UW60, Q8IY88, Q9UF79 | Gene names | PCSK4, PC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (Proprotein convertase PC4). | |||||
|
PCSK4_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.986491 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29121, Q62094 | Gene names | Pcsk4, Nec-3, Nec3 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (PC4) (Neuroendocrine convertase 3) (NEC 3) (Prohormone convertase 3) (KEX2-like endoprotease 3). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.745217 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
PCSK6_HUMAN
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.904758 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | 5.00115e-184 (rank : 7) | NC score | 0.892481 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
NEC1_HUMAN
|
||||||
| θ value | 4.26331e-159 (rank : 8) | NC score | 0.977727 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29120, P78478, Q92532 | Gene names | PCSK1, NEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroendocrine convertase 1 precursor (EC 3.4.21.93) (NEC 1) (PC1) (Prohormone convertase 1) (Proprotein convertase 1). | |||||
|
NEC1_MOUSE
|
||||||
| θ value | 1.24043e-158 (rank : 9) | NC score | 0.978216 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63239, P21662, P22546 | Gene names | Pcsk1, Att-1, Nec-1, Nec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine convertase 1 precursor (EC 3.4.21.93) (NEC 1) (PC1) (Prohormone convertase 1) (Proprotein convertase 1) (PC3) (Furin homolog) (Propeptide-processing protease). | |||||
|
NEC2_HUMAN
|
||||||
| θ value | 9.22056e-146 (rank : 10) | NC score | 0.975812 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16519, Q14927, Q9NQG3, Q9NUG1, Q9UJC6 | Gene names | PCSK2, NEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroendocrine convertase 2 precursor (EC 3.4.21.94) (NEC 2) (PC2) (Prohormone convertase 2) (Proprotein convertase 2) (KEX2-like endoprotease 2). | |||||
|
NEC2_MOUSE
|
||||||
| θ value | 6.60789e-144 (rank : 11) | NC score | 0.975830 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21661, Q80WU1 | Gene names | Pcsk2, Nec-2, Nec2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroendocrine convertase 2 precursor (EC 3.4.21.94) (NEC 2) (PC2) (Prohormone convertase 2) (Proprotein convertase 2) (KEX2-like endoprotease 2). | |||||
|
PCSK7_MOUSE
|
||||||
| θ value | 5.84277e-108 (rank : 12) | NC score | 0.961502 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61139, O08577 | Gene names | Pcsk7, Pc7 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 7 precursor (EC 3.4.21.-) (Proprotein convertase PC7) (Subtilisin/kexin-like protease PC7) (Prohormone convertase PC7) (Subtilisin-like proprotein convertase 7) (SPC7). | |||||
|
PCSK7_HUMAN
|
||||||
| θ value | 4.94618e-107 (rank : 13) | NC score | 0.961647 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16549, Q9UL57 | Gene names | PCSK7, LPC, PC7, PC8, SPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 7 precursor (EC 3.4.21.-) (Proprotein convertase PC7) (Subtilisin/kexin-like protease PC7) (Prohormone convertase PC7) (PC8) (hPC8) (Lymphoma proprotein convertase). | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 14) | NC score | 0.176082 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 15) | NC score | 0.174538 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
RSPO2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.174712 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BFU0, Q7TPX3 | Gene names | Rspo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (Cysteine-rich and single thrombospondin domain-containing protein 2) (Cristin-2) (mCristin-2). | |||||
|
RSPO3_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 17) | NC score | 0.178319 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BXY4, Q5VTV4, Q96K87 | Gene names | RSPO3, PWTSR, THSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (hRspo3) (Thrombospondin type-1 domain-containing protein 2) (Protein with TSP type-1 repeat) (hPWTSR). | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.080394 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
TNR1B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.063628 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
ERBB2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.041151 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
RSPO2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.171584 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UXX9, Q4G0U4, Q8N6X6 | Gene names | RSPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (hRspo2). | |||||
|
RSPO4_MOUSE
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.177752 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BJ73 | Gene names | Rspo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (Cysteine-rich and single thrombospondin domain-containing protein 4) (Cristin-4) (mCristin-4). | |||||
|
IGF1R_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.032966 (rank : 55) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
RSPO3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.175785 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2TJ95, Q3SYI9, Q5R2V4, Q8BVW2, Q9CSB2 | Gene names | Rspo3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (Cysteine-rich and single thrombospondin domain-containing protein 1) (Cristin-1) (Nucleopondin) (Cabriolet). | |||||
|
ERBB3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.046314 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
IGF1R_MOUSE
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.033717 (rank : 54) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60751, O70438, Q62123 | Gene names | Igf1r | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
SPG20_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.035578 (rank : 52) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0X7, O60349, Q86Y67, Q9H1T2, Q9H1T3 | Gene names | SPG20, KIAA0610, TAHCCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin (Trans-activated by hepatitis C virus core protein 1). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.089695 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.038929 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.063901 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
INSR_MOUSE
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.028930 (rank : 56) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 968 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15208 | Gene names | Insr | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor precursor (EC 2.7.10.1) (IR) (CD220 antigen) [Contains: Insulin receptor subunit alpha; Insulin receptor subunit beta]. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.090393 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
ERBB3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.045879 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
RSPO4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.170413 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
AXN2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.010176 (rank : 59) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
EGFR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.035329 (rank : 53) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
EGFR_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.036349 (rank : 51) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||
|
INSR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.027357 (rank : 57) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 963 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06213 | Gene names | INSR | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor precursor (EC 2.7.10.1) (IR) (CD220 antigen) [Contains: Insulin receptor subunit alpha; Insulin receptor subunit beta]. | |||||
|
RSPO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.163338 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q2MKA7, Q5T0F2, Q8N7L5 | Gene names | RSPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (hRspo1). | |||||
|
TNR11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.027065 (rank : 58) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
ERBB4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.040345 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
HECD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.005469 (rank : 60) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
BT2A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.005192 (rank : 61) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KV6 | Gene names | BTN2A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A3 precursor. | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.002287 (rank : 63) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
PCSK9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.056875 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NBP7, Q5PSM5 | Gene names | PCSK9, NARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 9 precursor (EC 3.4.21.-) (Proprotein convertase PC9) (Subtilisin/kexin-like protease PC9) (Neural apoptosis-regulated convertase 1) (NARC-1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | -0.001416 (rank : 64) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
GNDS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.003513 (rank : 62) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MBTP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.081478 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
MBTP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.075993 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTZ2, Q6PG67 | Gene names | Mbtps1, S1p, Ski1 | |||
|
Domain Architecture |
|
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin isozyme 1) (SKI-1) (Sterol-regulated luminal protease). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.061757 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.062716 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.061008 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.053111 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.059153 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.072104 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.067409 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052542 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.056535 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
RSPO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.148483 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z132, Q3V1S3 | Gene names | Rspo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (Cysteine-rich and single thrombospondin domain-containing protein 3) (Cristin-3) (mCristin-3). | |||||
|
TNR16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.055527 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
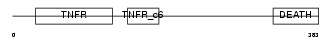
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050048 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |
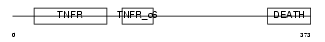
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
TNR1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.054475 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.054045 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TNR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.051020 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |
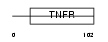
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
FURIN_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P23188 | Gene names | Furin, Fur, Pcsk3 | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme) (Prohormone convertase 3). | |||||
|
FURIN_HUMAN
|
||||||
| NC score | 0.998706 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P09958, Q14336, Q6LBS3 | Gene names | FURIN, FUR, PACE | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme). | |||||
|
PCSK4_HUMAN
|
||||||
| NC score | 0.990565 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6UW60, Q8IY88, Q9UF79 | Gene names | PCSK4, PC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (Proprotein convertase PC4). | |||||
|
PCSK4_MOUSE
|
||||||
| NC score | 0.986491 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P29121, Q62094 | Gene names | Pcsk4, Nec-3, Nec3 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (PC4) (Neuroendocrine convertase 3) (NEC 3) (Prohormone convertase 3) (KEX2-like endoprotease 3). | |||||
|
NEC1_MOUSE
|
||||||
| NC score | 0.978216 (rank : 5) | θ value | 1.24043e-158 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63239, P21662, P22546 | Gene names | Pcsk1, Att-1, Nec-1, Nec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine convertase 1 precursor (EC 3.4.21.93) (NEC 1) (PC1) (Prohormone convertase 1) (Proprotein convertase 1) (PC3) (Furin homolog) (Propeptide-processing protease). | |||||
|
NEC1_HUMAN
|
||||||
| NC score | 0.977727 (rank : 6) | θ value | 4.26331e-159 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P29120, P78478, Q92532 | Gene names | PCSK1, NEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroendocrine convertase 1 precursor (EC 3.4.21.93) (NEC 1) (PC1) (Prohormone convertase 1) (Proprotein convertase 1). | |||||
|
NEC2_MOUSE
|
||||||
| NC score | 0.975830 (rank : 7) | θ value | 6.60789e-144 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21661, Q80WU1 | Gene names | Pcsk2, Nec-2, Nec2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroendocrine convertase 2 precursor (EC 3.4.21.94) (NEC 2) (PC2) (Prohormone convertase 2) (Proprotein convertase 2) (KEX2-like endoprotease 2). | |||||
|
NEC2_HUMAN
|
||||||
| NC score | 0.975812 (rank : 8) | θ value | 9.22056e-146 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P16519, Q14927, Q9NQG3, Q9NUG1, Q9UJC6 | Gene names | PCSK2, NEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroendocrine convertase 2 precursor (EC 3.4.21.94) (NEC 2) (PC2) (Prohormone convertase 2) (Proprotein convertase 2) (KEX2-like endoprotease 2). | |||||
|
PCSK7_HUMAN
|
||||||
| NC score | 0.961647 (rank : 9) | θ value | 4.94618e-107 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16549, Q9UL57 | Gene names | PCSK7, LPC, PC7, PC8, SPC7 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 7 precursor (EC 3.4.21.-) (Proprotein convertase PC7) (Subtilisin/kexin-like protease PC7) (Prohormone convertase PC7) (PC8) (hPC8) (Lymphoma proprotein convertase). | |||||
|
PCSK7_MOUSE
|
||||||
| NC score | 0.961502 (rank : 10) | θ value | 5.84277e-108 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61139, O08577 | Gene names | Pcsk7, Pc7 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 7 precursor (EC 3.4.21.-) (Proprotein convertase PC7) (Subtilisin/kexin-like protease PC7) (Prohormone convertase PC7) (Subtilisin-like proprotein convertase 7) (SPC7). | |||||
|
PCSK6_HUMAN
|
||||||
| NC score | 0.904758 (rank : 11) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.892481 (rank : 12) | θ value | 5.00115e-184 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.745217 (rank : 13) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
RSPO3_HUMAN
|
||||||
| NC score | 0.178319 (rank : 14) | θ value | 0.0736092 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9BXY4, Q5VTV4, Q96K87 | Gene names | RSPO3, PWTSR, THSD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (hRspo3) (Thrombospondin type-1 domain-containing protein 2) (Protein with TSP type-1 repeat) (hPWTSR). | |||||
|
RSPO4_MOUSE
|
||||||
| NC score | 0.177752 (rank : 15) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BJ73 | Gene names | Rspo4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (Cysteine-rich and single thrombospondin domain-containing protein 4) (Cristin-4) (mCristin-4). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.176082 (rank : 16) | θ value | 1.53129e-07 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
RSPO3_MOUSE
|
||||||
| NC score | 0.175785 (rank : 17) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2TJ95, Q3SYI9, Q5R2V4, Q8BVW2, Q9CSB2 | Gene names | Rspo3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-3 precursor (Roof plate-specific spondin-3) (Cysteine-rich and single thrombospondin domain-containing protein 1) (Cristin-1) (Nucleopondin) (Cabriolet). | |||||
|
RSPO2_MOUSE
|
||||||
| NC score | 0.174712 (rank : 18) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BFU0, Q7TPX3 | Gene names | Rspo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (Cysteine-rich and single thrombospondin domain-containing protein 2) (Cristin-2) (mCristin-2). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.174538 (rank : 19) | θ value | 2.88788e-06 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
RSPO2_HUMAN
|
||||||
| NC score | 0.171584 (rank : 20) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6UXX9, Q4G0U4, Q8N6X6 | Gene names | RSPO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-2 precursor (Roof plate-specific spondin-2) (hRspo2). | |||||
|
RSPO4_HUMAN
|
||||||
| NC score | 0.170413 (rank : 21) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
RSPO1_HUMAN
|
||||||
| NC score | 0.163338 (rank : 22) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q2MKA7, Q5T0F2, Q8N7L5 | Gene names | RSPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (hRspo1). | |||||
|
RSPO1_MOUSE
|
||||||
| NC score | 0.148483 (rank : 23) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Z132, Q3V1S3 | Gene names | Rspo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (Cysteine-rich and single thrombospondin domain-containing protein 3) (Cristin-3) (mCristin-3). | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.090393 (rank : 24) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.089695 (rank : 25) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
MBTP1_HUMAN
|
||||||
| NC score | 0.081478 (rank : 26) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.080394 (rank : 27) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
MBTP1_MOUSE
|
||||||
| NC score | 0.075993 (rank : 28) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WTZ2, Q6PG67 | Gene names | Mbtps1, S1p, Ski1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin isozyme 1) (SKI-1) (Sterol-regulated luminal protease). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.072104 (rank : 29) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.067409 (rank : 30) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.063901 (rank : 31) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TNR1B_HUMAN
|
||||||
| NC score | 0.063628 (rank : 32) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.062716 (rank : 33) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.061757 (rank : 34) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.061008 (rank : 35) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.059153 (rank : 36) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
PCSK9_HUMAN
|
||||||
| NC score | 0.056875 (rank : 37) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NBP7, Q5PSM5 | Gene names | PCSK9, NARC1 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 9 precursor (EC 3.4.21.-) (Proprotein convertase PC9) (Subtilisin/kexin-like protease PC9) (Neural apoptosis-regulated convertase 1) (NARC-1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.056535 (rank : 38) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.055527 (rank : 39) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR1A_MOUSE
|
||||||
| NC score | 0.054475 (rank : 40) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.054045 (rank : 41) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.053111 (rank : 42) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.052542 (rank : 43) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
TNR9_MOUSE
|
||||||
| NC score | 0.051020 (rank : 44) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
TNR16_MOUSE
|
||||||
| NC score | 0.050048 (rank : 45) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
ERBB3_MOUSE
|
||||||
| NC score | 0.046314 (rank : 46) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
ERBB3_HUMAN
|
||||||
| NC score | 0.045879 (rank : 47) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
ERBB2_MOUSE
|
||||||
| NC score | 0.041151 (rank : 48) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
ERBB4_HUMAN
|
||||||
| NC score | 0.040345 (rank : 49) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 910 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q15303 | Gene names | ERBB4, HER4 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-4 precursor (EC 2.7.10.1) (p180erbB4) (Tyrosine kinase-type cell surface receptor HER4). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.038929 (rank : 50) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
EGFR_MOUSE
|
||||||
| NC score | 0.036349 (rank : 51) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||
|
SPG20_HUMAN
|
||||||
| NC score | 0.035578 (rank : 52) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N0X7, O60349, Q86Y67, Q9H1T2, Q9H1T3 | Gene names | SPG20, KIAA0610, TAHCCP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spartin (Trans-activated by hepatitis C virus core protein 1). | |||||
|
EGFR_HUMAN
|
||||||
| NC score | 0.035329 (rank : 53) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
IGF1R_MOUSE
|
||||||
| NC score | 0.033717 (rank : 54) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q60751, O70438, Q62123 | Gene names | Igf1r | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
IGF1R_HUMAN
|
||||||
| NC score | 0.032966 (rank : 55) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
INSR_MOUSE
|
||||||
| NC score | 0.028930 (rank : 56) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 968 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P15208 | Gene names | Insr | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor precursor (EC 2.7.10.1) (IR) (CD220 antigen) [Contains: Insulin receptor subunit alpha; Insulin receptor subunit beta]. | |||||
|
INSR_HUMAN
|
||||||
| NC score | 0.027357 (rank : 57) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 963 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06213 | Gene names | INSR | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor precursor (EC 2.7.10.1) (IR) (CD220 antigen) [Contains: Insulin receptor subunit alpha; Insulin receptor subunit beta]. | |||||
|
TNR11_MOUSE
|
||||||
| NC score | 0.027065 (rank : 58) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
AXN2_MOUSE
|
||||||
| NC score | 0.010176 (rank : 59) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88566, Q9QXJ6 | Gene names | Axin2 | |||
|
Domain Architecture |

|
|||||
| Description | Axin-2 (Axis inhibition protein 2) (Conductin) (Axin-like protein) (Axil). | |||||
|
HECD1_HUMAN
|
||||||
| NC score | 0.005469 (rank : 60) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULT8, Q6P445, Q86VJ1, Q96F34, Q9UFZ7 | Gene names | HECTD1, KIAA1131 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase HECTD1 (HECT domain-containing protein 1) (E3 ligase for inhibin receptor) (EULIR). | |||||
|
BT2A3_HUMAN
|
||||||
| NC score | 0.005192 (rank : 61) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KV6 | Gene names | BTN2A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Butyrophilin subfamily 2 member A3 precursor. | |||||
|
GNDS_MOUSE
|
||||||
| NC score | 0.003513 (rank : 62) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q03385 | Gene names | Ralgds, Rgds | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.002287 (rank : 63) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.001416 (rank : 64) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||