Please be patient as the page loads
|
TRAF1_HUMAN
|
||||||
| SwissProt Accessions | Q13077, Q8NF13 | Gene names | TRAF1, EBI6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 1 (Epstein-Barr virus-induced protein 6). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TRAF1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13077, Q8NF13 | Gene names | TRAF1, EBI6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 1 (Epstein-Barr virus-induced protein 6). | |||||
|
TRAF1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993061 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39428 | Gene names | Traf1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 1. | |||||
|
TRAF2_HUMAN
|
||||||
| θ value | 1.22964e-73 (rank : 3) | NC score | 0.865942 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12933, Q96NT2 | Gene names | TRAF2, TRAP3 | |||
|
Domain Architecture |
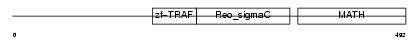
|
|||||
| Description | TNF receptor-associated factor 2 (Tumor necrosis factor type 2 receptor-associated protein 3). | |||||
|
TRAF2_MOUSE
|
||||||
| θ value | 3.57772e-73 (rank : 4) | NC score | 0.859397 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39429, O54896 | Gene names | Traf2 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2. | |||||
|
TRAF3_MOUSE
|
||||||
| θ value | 5.35859e-61 (rank : 5) | NC score | 0.808803 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60803, Q62380 | Gene names | Traf3, Craf1, Trafamn | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (TRAFAMN). | |||||
|
TRAF3_HUMAN
|
||||||
| θ value | 1.55911e-60 (rank : 6) | NC score | 0.808186 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13114, Q12990, Q13076, Q13947, Q6AZX1, Q9UNL1 | Gene names | TRAF3, CAP1, CRAF1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (CD40-binding protein) (CD40BP) (LMP1-associated protein) (LAP1) (CAP-1). | |||||
|
TRAF5_MOUSE
|
||||||
| θ value | 4.39379e-55 (rank : 7) | NC score | 0.811560 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70191, Q61480 | Gene names | Traf5 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5. | |||||
|
TRAF5_HUMAN
|
||||||
| θ value | 2.18062e-54 (rank : 8) | NC score | 0.812956 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
TRAF4_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 9) | NC score | 0.711458 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BUZ4, O75615, Q14848, Q2PJN8 | Gene names | TRAF4, CART1, MLN62, RNF83 | |||
|
Domain Architecture |
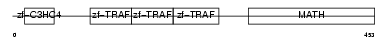
|
|||||
| Description | TNF receptor-associated factor 4 (Cysteine-rich domain associated with RING and Traf domains protein 1) (Malignant 62) (RING finger protein 83). | |||||
|
TRAF4_MOUSE
|
||||||
| θ value | 5.97985e-28 (rank : 10) | NC score | 0.711175 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61382 | Gene names | Traf4, Cart1 | |||
|
Domain Architecture |
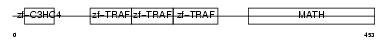
|
|||||
| Description | TNF receptor-associated factor 4 (Cysteine-rich motif associated to RING and Traf domains protein 1). | |||||
|
TRAF6_HUMAN
|
||||||
| θ value | 1.6852e-22 (rank : 11) | NC score | 0.698796 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y4K3, Q8NEH5 | Gene names | TRAF6, RNF85 | |||
|
Domain Architecture |
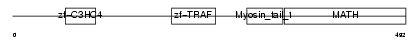
|
|||||
| Description | TNF receptor-associated factor 6 (Interleukin 1 signal transducer) (RING finger protein 85). | |||||
|
TRAF6_MOUSE
|
||||||
| θ value | 1.86321e-21 (rank : 12) | NC score | 0.697186 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70196, Q8BLV2 | Gene names | Traf6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 6. | |||||
|
MEP1A_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 13) | NC score | 0.215192 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28825 | Gene names | Mep1a | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (MEP-1). | |||||
|
MEP1A_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.182525 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16819 | Gene names | MEP1A | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit alpha) (PABA peptide hydrolase) (PPH alpha). | |||||
|
MEP1B_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.216991 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61847 | Gene names | Mep1b, Mep-1b | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.044060 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.040440 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.044003 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.046002 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.054608 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.055262 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SIAH2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.060308 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43255, O43270 | Gene names | SIAH2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in absentia homolog 2) (Siah-2) (hSiah2). | |||||
|
SIAH2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.060301 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06986 | Gene names | Siah2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in absentia homolog 2) (Siah-2) (mSiah2). | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.064489 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.041372 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.018526 (rank : 117) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
CKAP4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.048209 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.043963 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.050795 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.030600 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.012918 (rank : 120) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.018734 (rank : 116) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.042195 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
NIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.041084 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
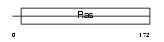
RAB6C_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.006112 (rank : 124) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0N0, Q9P128 | Gene names | RAB6C, WTH3 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-6C (Rab6-like protein WTH3). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.020732 (rank : 115) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.024734 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
RUFY2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.035412 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
CCD62_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.034607 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.044215 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.052908 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.017767 (rank : 118) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.029589 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.039795 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
COG7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.022584 (rank : 112) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P83436, Q6UWU7 | Gene names | COG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Conserved oligomeric Golgi complex component 7. | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.011699 (rank : 121) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.022202 (rank : 113) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.044367 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.034637 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.038478 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MYH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.044096 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
SYTL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.005287 (rank : 125) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.022701 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.029124 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.039194 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.017110 (rank : 119) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
EMIL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.007694 (rank : 123) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.040758 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
HOOK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.027654 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
I20RA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.007940 (rank : 122) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHF4, Q6UWA9, Q96SH8 | Gene names | IL20RA, ZCYTOR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-20 receptor alpha chain precursor (IL-20R-alpha) (IL-20R1) (Cytokine receptor family 2 member 8) (Cytokine receptor class-II member 8) (CRF2-8) (ZcytoR7). | |||||
|
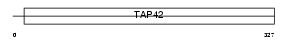
IGBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.020963 (rank : 114) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61249 | Gene names | Igbp1, Pc52 | |||
|
Domain Architecture |
|
|||||
| Description | Immunoglobulin-binding protein 1 (CD79a-binding protein 1) (Alpha4 phosphoprotein) (Lymphocyte signal transduction molecule alpha 4) (p52). | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.036718 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.042332 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.037600 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.032404 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
BFAR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.077128 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
BFAR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.072971 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.073350 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.070982 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
CEP57_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.052731 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
CKAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.050540 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.052441 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.052324 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
FBX28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.055130 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVF7, O75070 | Gene names | FBXO28, KIAA0483 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 28. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050486 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.052625 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
LNX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.083908 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
LNX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.084288 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70263, O70264, Q8BRI8, Q8CFR3 | Gene names | Lnx1, Lnx | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
LNX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.065556 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N448, Q5W0P0, Q6ZMH2, Q96SH4 | Gene names | LNX2, PDZRN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2) (PDZ domain- containing RING finger protein 1). | |||||
|
LNX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.069535 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XL2, Q8CBE1 | Gene names | Lnx2 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2). | |||||
|
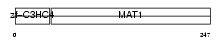
MAT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.054040 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51948, Q15817 | Gene names | MNAT1, CAP35, MAT1, RNF66 | |||
|
Domain Architecture |
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35) (Cyclin G1-interacting protein) (RING finger protein 66). | |||||
|
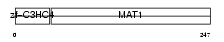
MAT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.054979 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
MEP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.176297 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16820, Q670J1 | Gene names | MEP1B | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit beta) (PABA peptide hydrolase) (PPH beta). | |||||
|
PEX10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.066260 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |
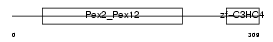
|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.141510 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.144682 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.108543 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
R113A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.082689 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
R113B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.072534 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
RAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.104503 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15918, Q8NER2 | Gene names | RAG1, RNF74 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1) (RING finger protein 74). | |||||
|
RAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.105237 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15919 | Gene names | Rag1, Rag-1 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1). | |||||
|
RING1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.095400 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |
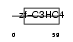
|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.093554 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
RING2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.085388 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99496, Q5TEN1, Q5TEN2 | Gene names | RNF2, BAP1, DING, HIPI3, RING1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein RING2 (EC 6.3.2.-) (RING finger protein 2) (RING finger protein 1B) (RING1b) (RING finger protein BAP-1) (DinG protein) (Huntingtin-interacting protein 2-interacting protein 3) (HIP2-interacting protein 3). | |||||
|
RING2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.085379 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQJ4, O35699, O35729, Q4FJV5, Q8C1X8 | Gene names | Rnf2, DinG, Ring1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein RING2 (EC 6.3.2.-) (RING finger protein 2) (RING finger protein 1B) (RING1b). | |||||
|
RN125_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.164769 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EQ8, Q9NX39 | Gene names | RNF125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-) (T-cell RING activation protein 1) (TRAC-1). | |||||
|
RN125_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.138552 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
RN166_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.193016 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96A37, Q96DM0 | Gene names | RNF166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
RN166_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.187645 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U9F6 | Gene names | Rnf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
RNF41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.164391 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H4P4, O75598 | Gene names | RNF41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 41. | |||||
|
RNF41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.163506 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH75, Q8BGJ2, Q8BMC9, Q8VEA2, Q9CRK6, Q9D568 | Gene names | Rnf41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 41. | |||||
|
RNF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.060283 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78317 | Gene names | RNF4 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 4. | |||||
|
RNF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.063090 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZS2, O35941, Q541Z6 | Gene names | Rnf4 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 4. | |||||
|
TRAF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.137336 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6Q0C0, Q9H073 | Gene names | TRAF7, RFWD1, RNF119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-) (TNF receptor- associated factor 7) (RING finger and WD repeat domain protein 1) (RING finger protein 119). | |||||
|
TRAF7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.139611 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q922B6, Q99JV3 | Gene names | Traf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-) (TNF receptor- associated factor 7). | |||||
|
TRI12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.060159 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PQ1, Q9D704 | Gene names | Trim12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 12. | |||||
|
TRI22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053123 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IYM9, Q15521 | Gene names | TRIM22, RNF94, STAF50 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 22 (RING finger protein 94) (50 kDa-stimulated trans-acting factor) (Staf-50). | |||||
|
TRI25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.056154 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
TRI25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.063439 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61510 | Gene names | Trim25, Efp, Zfp147, Znf147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp). | |||||
|
TRI26_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.053980 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12899 | Gene names | TRIM26, RNF95, ZNF173 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 26 (Zinc finger protein 173) (Acid finger protein) (AFP) (RING finger protein 95). | |||||
|
TRI26_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.052317 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PN3, Q8C9E9, Q8JZT7, Q99PN2 | Gene names | Trim26, Znf173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 26 (Zinc finger protein 173). | |||||
|
TRI31_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.071780 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZY9, Q53H52, Q5RI37, Q5SRJ7, Q5SRJ8, Q5SS28, Q96AK4, Q96AP8, Q99579, Q9BZY8 | Gene names | TRIM31, C6orf13, RNF | |||
|
Domain Architecture |
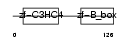
|
|||||
| Description | Tripartite motif-containing protein 31. | |||||
|
TRI47_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.058377 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
TRI47_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.067355 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
TRI56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.053920 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
TRI65_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.055295 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PJ69, Q4G0F0, Q6DKJ6, Q9BRP6 | Gene names | TRIM65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 65. | |||||
|
TRI73_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.055940 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UV7, Q8N0S3 | Gene names | TRIM73, TRIM50B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 73 (Tripartite motif-containing protein 50B). | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.056647 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.057202 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.060831 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.060104 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.079035 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.077403 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
ZN313_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.147353 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y508, Q6N0B0 | Gene names | ZNF313, RNF114, ZNF228 | |||
|
Domain Architecture |
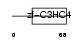
|
|||||
| Description | Zinc finger protein 313 (RING finger protein 114). | |||||
|
ZN313_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.154475 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ET26, Q3UFU8, Q8K5A2 | Gene names | Znf313, Zfp228, Zfp313, Znf228 | |||
|
Domain Architecture |
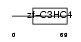
|
|||||
| Description | Zinc finger protein 313. | |||||
|
TRAF1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q13077, Q8NF13 | Gene names | TRAF1, EBI6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 1 (Epstein-Barr virus-induced protein 6). | |||||
|
TRAF1_MOUSE
|
||||||
| NC score | 0.993061 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P39428 | Gene names | Traf1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 1. | |||||
|
TRAF2_HUMAN
|
||||||
| NC score | 0.865942 (rank : 3) | θ value | 1.22964e-73 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q12933, Q96NT2 | Gene names | TRAF2, TRAP3 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2 (Tumor necrosis factor type 2 receptor-associated protein 3). | |||||
|
TRAF2_MOUSE
|
||||||
| NC score | 0.859397 (rank : 4) | θ value | 3.57772e-73 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39429, O54896 | Gene names | Traf2 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 2. | |||||
|
TRAF5_HUMAN
|
||||||
| NC score | 0.812956 (rank : 5) | θ value | 2.18062e-54 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00463 | Gene names | TRAF5, RNF84 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5 (RING finger protein 84). | |||||
|
TRAF5_MOUSE
|
||||||
| NC score | 0.811560 (rank : 6) | θ value | 4.39379e-55 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70191, Q61480 | Gene names | Traf5 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 5. | |||||
|
TRAF3_MOUSE
|
||||||
| NC score | 0.808803 (rank : 7) | θ value | 5.35859e-61 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q60803, Q62380 | Gene names | Traf3, Craf1, Trafamn | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (TRAFAMN). | |||||
|
TRAF3_HUMAN
|
||||||
| NC score | 0.808186 (rank : 8) | θ value | 1.55911e-60 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q13114, Q12990, Q13076, Q13947, Q6AZX1, Q9UNL1 | Gene names | TRAF3, CAP1, CRAF1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 3 (CD40 receptor-associated factor 1) (CRAF1) (CD40-binding protein) (CD40BP) (LMP1-associated protein) (LAP1) (CAP-1). | |||||
|
TRAF4_HUMAN
|
||||||
| NC score | 0.711458 (rank : 9) | θ value | 1.85889e-29 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 373 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BUZ4, O75615, Q14848, Q2PJN8 | Gene names | TRAF4, CART1, MLN62, RNF83 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 4 (Cysteine-rich domain associated with RING and Traf domains protein 1) (Malignant 62) (RING finger protein 83). | |||||
|
TRAF4_MOUSE
|
||||||
| NC score | 0.711175 (rank : 10) | θ value | 5.97985e-28 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61382 | Gene names | Traf4, Cart1 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 4 (Cysteine-rich motif associated to RING and Traf domains protein 1). | |||||
|
TRAF6_HUMAN
|
||||||
| NC score | 0.698796 (rank : 11) | θ value | 1.6852e-22 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y4K3, Q8NEH5 | Gene names | TRAF6, RNF85 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 6 (Interleukin 1 signal transducer) (RING finger protein 85). | |||||
|
TRAF6_MOUSE
|
||||||
| NC score | 0.697186 (rank : 12) | θ value | 1.86321e-21 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70196, Q8BLV2 | Gene names | Traf6 | |||
|
Domain Architecture |

|
|||||
| Description | TNF receptor-associated factor 6. | |||||
|
MEP1B_MOUSE
|
||||||
| NC score | 0.216991 (rank : 13) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61847 | Gene names | Mep1b, Mep-1b | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2). | |||||
|
MEP1A_MOUSE
|
||||||
| NC score | 0.215192 (rank : 14) | θ value | 0.00020696 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P28825 | Gene names | Mep1a | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (MEP-1). | |||||
|
RN166_HUMAN
|
||||||
| NC score | 0.193016 (rank : 15) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96A37, Q96DM0 | Gene names | RNF166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
RN166_MOUSE
|
||||||
| NC score | 0.187645 (rank : 16) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3U9F6 | Gene names | Rnf166 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 166. | |||||
|
MEP1A_HUMAN
|
||||||
| NC score | 0.182525 (rank : 17) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16819 | Gene names | MEP1A | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit alpha precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit alpha) (PABA peptide hydrolase) (PPH alpha). | |||||
|
MEP1B_HUMAN
|
||||||
| NC score | 0.176297 (rank : 18) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16820, Q670J1 | Gene names | MEP1B | |||
|
Domain Architecture |

|
|||||
| Description | Meprin A subunit beta precursor (EC 3.4.24.18) (Endopeptidase-2) (N- benzoyl-L-tyrosyl-P-amino-benzoic acid hydrolase subunit beta) (PABA peptide hydrolase) (PPH beta). | |||||
|
RN125_HUMAN
|
||||||
| NC score | 0.164769 (rank : 19) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96EQ8, Q9NX39 | Gene names | RNF125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-) (T-cell RING activation protein 1) (TRAC-1). | |||||
|
RNF41_HUMAN
|
||||||
| NC score | 0.164391 (rank : 20) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H4P4, O75598 | Gene names | RNF41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 41. | |||||
|
RNF41_MOUSE
|
||||||
| NC score | 0.163506 (rank : 21) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BH75, Q8BGJ2, Q8BMC9, Q8VEA2, Q9CRK6, Q9D568 | Gene names | Rnf41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 41. | |||||
|
ZN313_MOUSE
|
||||||
| NC score | 0.154475 (rank : 22) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ET26, Q3UFU8, Q8K5A2 | Gene names | Znf313, Zfp228, Zfp313, Znf228 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 313. | |||||
|
ZN313_HUMAN
|
||||||
| NC score | 0.147353 (rank : 23) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y508, Q6N0B0 | Gene names | ZNF313, RNF114, ZNF228 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 313 (RING finger protein 114). | |||||
|
PZRN3_MOUSE
|
||||||
| NC score | 0.144682 (rank : 24) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q69ZS0, Q91Z03, Q9QY54, Q9QY55 | Gene names | Pdzrn3, Kiaa1095, Semcap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.141510 (rank : 25) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
TRAF7_MOUSE
|
||||||
| NC score | 0.139611 (rank : 26) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q922B6, Q99JV3 | Gene names | Traf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-) (TNF receptor- associated factor 7). | |||||
|
RN125_MOUSE
|
||||||
| NC score | 0.138552 (rank : 27) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
TRAF7_HUMAN
|
||||||
| NC score | 0.137336 (rank : 28) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6Q0C0, Q9H073 | Gene names | TRAF7, RFWD1, RNF119 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin protein ligase TRAF7 (EC 6.3.2.-) (TNF receptor- associated factor 7) (RING finger and WD repeat domain protein 1) (RING finger protein 119). | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.108543 (rank : 29) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
RAG1_MOUSE
|
||||||
| NC score | 0.105237 (rank : 30) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15919 | Gene names | Rag1, Rag-1 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1). | |||||
|
RAG1_HUMAN
|
||||||
| NC score | 0.104503 (rank : 31) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P15918, Q8NER2 | Gene names | RAG1, RNF74 | |||
|
Domain Architecture |

|
|||||
| Description | V(D)J recombination-activating protein 1 (RAG-1) (RING finger protein 74). | |||||
|
RING1_HUMAN
|
||||||
| NC score | 0.095400 (rank : 32) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.093554 (rank : 33) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
RING2_HUMAN
|
||||||
| NC score | 0.085388 (rank : 34) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99496, Q5TEN1, Q5TEN2 | Gene names | RNF2, BAP1, DING, HIPI3, RING1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein RING2 (EC 6.3.2.-) (RING finger protein 2) (RING finger protein 1B) (RING1b) (RING finger protein BAP-1) (DinG protein) (Huntingtin-interacting protein 2-interacting protein 3) (HIP2-interacting protein 3). | |||||
|
RING2_MOUSE
|
||||||
| NC score | 0.085379 (rank : 35) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQJ4, O35699, O35729, Q4FJV5, Q8C1X8 | Gene names | Rnf2, DinG, Ring1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein RING2 (EC 6.3.2.-) (RING finger protein 2) (RING finger protein 1B) (RING1b). | |||||
|
LNX1_MOUSE
|
||||||
| NC score | 0.084288 (rank : 36) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O70263, O70264, Q8BRI8, Q8CFR3 | Gene names | Lnx1, Lnx | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
LNX1_HUMAN
|
||||||
| NC score | 0.083908 (rank : 37) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TBB1, Q8N4C2, Q96MJ7, Q9BY20 | Gene names | LNX1, LNX | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase LNX (EC 6.3.2.-) (Numb-binding protein 1) (Ligand of Numb-protein X 1). | |||||
|
R113A_HUMAN
|
||||||
| NC score | 0.082689 (rank : 38) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15541 | Gene names | RNF113A, RNF113, ZNF183 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113A (Zinc finger protein 183). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.079035 (rank : 39) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.077403 (rank : 40) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
BFAR_HUMAN
|
||||||
| NC score | 0.077128 (rank : 41) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NZS9 | Gene names | BFAR, BAR, RNF47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator (RING finger protein 47). | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.073350 (rank : 42) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BFAR_MOUSE
|
||||||
| NC score | 0.072971 (rank : 43) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R079, Q8C1A7, Q9CXY3 | Gene names | Bfar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bifunctional apoptosis regulator. | |||||
|
R113B_HUMAN
|
||||||
| NC score | 0.072534 (rank : 44) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IZP6, Q8WWF9, Q96QY9 | Gene names | RNF113B, RNF161, ZNF183L1 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 113B (Zinc finger protein 183-like 1). | |||||
|
TRI31_HUMAN
|
||||||
| NC score | 0.071780 (rank : 45) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BZY9, Q53H52, Q5RI37, Q5SRJ7, Q5SRJ8, Q5SS28, Q96AK4, Q96AP8, Q99579, Q9BZY8 | Gene names | TRIM31, C6orf13, RNF | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 31. | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.070982 (rank : 46) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
LNX2_MOUSE
|
||||||
| NC score | 0.069535 (rank : 47) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XL2, Q8CBE1 | Gene names | Lnx2 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2). | |||||
|
TRI47_MOUSE
|
||||||
| NC score | 0.067355 (rank : 48) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8C0E3, Q6P249, Q811J7, Q8BVZ8, Q8R1K0, Q8R3Y1 | Gene names | Trim47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47. | |||||
|
PEX10_HUMAN
|
||||||
| NC score | 0.066260 (rank : 49) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60683, Q5T095, Q9BW90 | Gene names | PEX10, RNF69 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 10 (Peroxin-10) (Peroxisome biogenesis factor 10) (RING finger protein 69). | |||||
|
LNX2_HUMAN
|
||||||
| NC score | 0.065556 (rank : 50) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N448, Q5W0P0, Q6ZMH2, Q96SH4 | Gene names | LNX2, PDZRN1 | |||
|
Domain Architecture |

|
|||||
| Description | Ligand of Numb-protein X 2 (Numb-binding protein 2) (PDZ domain- containing RING finger protein 1). | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.064489 (rank : 51) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
TRI25_MOUSE
|
||||||
| NC score | 0.063439 (rank : 52) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q61510 | Gene names | Trim25, Efp, Zfp147, Znf147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp). | |||||
|
RNF4_MOUSE
|
||||||
| NC score | 0.063090 (rank : 53) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QZS2, O35941, Q541Z6 | Gene names | Rnf4 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 4. | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.060831 (rank : 54) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
SIAH2_HUMAN
|
||||||
| NC score | 0.060308 (rank : 55) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43255, O43270 | Gene names | SIAH2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in absentia homolog 2) (Siah-2) (hSiah2). | |||||
|
SIAH2_MOUSE
|
||||||
| NC score | 0.060301 (rank : 56) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q06986 | Gene names | Siah2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase SIAH2 (EC 6.3.2.-) (Seven in absentia homolog 2) (Siah-2) (mSiah2). | |||||
|
RNF4_HUMAN
|
||||||
| NC score | 0.060283 (rank : 57) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P78317 | Gene names | RNF4 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 4. | |||||
|
TRI12_MOUSE
|
||||||
| NC score | 0.060159 (rank : 58) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q99PQ1, Q9D704 | Gene names | Trim12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 12. | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.060104 (rank : 59) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
TRI47_HUMAN
|
||||||
| NC score | 0.058377 (rank : 60) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96LD4, Q96GU5, Q9BRN7 | Gene names | TRIM47, GOA, RNF100 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 47 (Gene overexpressed in astrocytoma protein) (RING finger protein 100). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.057202 (rank : 61) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.056647 (rank : 62) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
TRI25_HUMAN
|
||||||
| NC score | 0.056154 (rank : 63) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14258 | Gene names | TRIM25, EFP, RNF147, ZNF147 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 25 (Zinc finger protein 147) (Estrogen-responsive finger protein) (Efp) (RING finger protein 147). | |||||
|
TRI73_HUMAN
|
||||||
| NC score | 0.055940 (rank : 64) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86UV7, Q8N0S3 | Gene names | TRIM73, TRIM50B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 73 (Tripartite motif-containing protein 50B). | |||||
|
TRI65_HUMAN
|
||||||
| NC score | 0.055295 (rank : 65) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6PJ69, Q4G0F0, Q6DKJ6, Q9BRP6 | Gene names | TRIM65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 65. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.055262 (rank : 66) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
FBX28_HUMAN
|
||||||
| NC score | 0.055130 (rank : 67) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NVF7, O75070 | Gene names | FBXO28, KIAA0483 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 28. | |||||
|
MAT1_MOUSE
|
||||||
| NC score | 0.054979 (rank : 68) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.054608 (rank : 69) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MAT1_HUMAN
|
||||||
| NC score | 0.054040 (rank : 70) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51948, Q15817 | Gene names | MNAT1, CAP35, MAT1, RNF66 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35) (Cyclin G1-interacting protein) (RING finger protein 66). | |||||
|
TRI26_HUMAN
|
||||||
| NC score | 0.053980 (rank : 71) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12899 | Gene names | TRIM26, RNF95, ZNF173 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 26 (Zinc finger protein 173) (Acid finger protein) (AFP) (RING finger protein 95). | |||||
|
TRI56_HUMAN
|
||||||
| NC score | 0.053920 (rank : 72) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BRZ2, Q86VT6, Q8N2H8, Q8NAC0, Q9H031 | Gene names | TRIM56, RNF109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 56 (RING finger protein 109). | |||||
|
TRI22_HUMAN
|
||||||
| NC score | 0.053123 (rank : 73) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IYM9, Q15521 | Gene names | TRIM22, RNF94, STAF50 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 22 (RING finger protein 94) (50 kDa-stimulated trans-acting factor) (Staf-50). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.052908 (rank : 74) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
CEP57_MOUSE
|
||||||
| NC score | 0.052731 (rank : 75) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 580 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CEE0, Q6ZQJ3, Q7TN18, Q80X65, Q810F2, Q9D4J4, Q9D5S4, Q9D5W5 | Gene names | Cep57, Kiaa0092, Tsp57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 57 kDa (Cep57 protein) (Testis-specific protein 57) (Translokin). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.052625 (rank : 76) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.052441 (rank : 77) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.052324 (rank : 78) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
TRI26_MOUSE
|
||||||
| NC score | 0.052317 (rank : 79) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99PN3, Q8C9E9, Q8JZT7, Q99PN2 | Gene names | Trim26, Znf173 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 26 (Zinc finger protein 173). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.050795 (rank : 80) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CKAP4_HUMAN
|
||||||
| NC score | 0.050540 (rank : 81) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07065, Q504S5, Q53ES6 | Gene names | CKAP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.050486 (rank : 82) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
CKAP4_MOUSE
|
||||||
| NC score | 0.048209 (rank : 83) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BMK4, Q8BTK8, Q8R3F2 | Gene names | Ckap4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-associated protein 4 (63 kDa membrane protein) (p63). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.046002 (rank : 84) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.044367 (rank : 85) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.044215 (rank : 86) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.044096 (rank : 87) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.044060 (rank : 88) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.044003 (rank : 89) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.043963 (rank : 90) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.042332 (rank : 91) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.042195 (rank : 92) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.041372 (rank : 93) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.041084 (rank : 94) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.040758 (rank : 95) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.040440 (rank : 96) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.039795 (rank : 97) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.039194 (rank : 98) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.038478 (rank : 99) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.037600 (rank : 100) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.036718 (rank : 101) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
RUFY2_MOUSE
|
||||||
| NC score | 0.035412 (rank : 102) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4C2, Q69ZH1 | Gene names | Rufy2, Kiaa1537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Leucine zipper FYVE-finger protein) (LZ-FYVE). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.034637 (rank : 103) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.034607 (rank : 104) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.032404 (rank : 105) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.030600 (rank : 106) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.029589 (rank : 107) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.029124 (rank : 108) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
HOOK2_HUMAN
|
||||||
| NC score | 0.027654 (rank : 109) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96ED9, O60562 | Gene names | HOOK2 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 2 (h-hook2) (hHK2). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.024734 (rank : 110) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.022701 (rank : 111) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
COG7_HUMAN
|
||||||
| NC score | 0.022584 (rank : 112) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P83436, Q6UWU7 | Gene names | COG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Conserved oligomeric Golgi complex component 7. | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.022202 (rank : 113) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
IGBP1_MOUSE
|
||||||
| NC score | 0.020963 (rank : 114) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61249 | Gene names | Igbp1, Pc52 | |||
|
Domain Architecture |

|
|||||
| Description | Immunoglobulin-binding protein 1 (CD79a-binding protein 1) (Alpha4 phosphoprotein) (Lymphocyte signal transduction molecule alpha 4) (p52). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.020732 (rank : 115) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.018734 (rank : 116) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.018526 (rank : 117) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.017767 (rank : 118) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.017110 (rank : 119) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.012918 (rank : 120) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.011699 (rank : 121) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
I20RA_HUMAN
|
||||||
| NC score | 0.007940 (rank : 122) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UHF4, Q6UWA9, Q96SH8 | Gene names | IL20RA, ZCYTOR7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-20 receptor alpha chain precursor (IL-20R-alpha) (IL-20R1) (Cytokine receptor family 2 member 8) (Cytokine receptor class-II member 8) (CRF2-8) (ZcytoR7). | |||||
|
EMIL1_HUMAN
|
||||||
| NC score | 0.007694 (rank : 123) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y6C2, Q96G58, Q9UG76 | Gene names | EMILIN1, EMI | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-1 precursor (Elastin microfibril interface-located protein 1) (Elastin microfibril interfacer 1). | |||||
|
RAB6C_HUMAN
|
||||||
| NC score | 0.006112 (rank : 124) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H0N0, Q9P128 | Gene names | RAB6C, WTH3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-6C (Rab6-like protein WTH3). | |||||
|
SYTL1_HUMAN
|
||||||
| NC score | 0.005287 (rank : 125) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYJ3, Q96BB6, Q96GU6, Q96S89, Q96SI0 | Gene names | SYTL1, SLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7) (Protein JFC1) (SB146). | |||||