Please be patient as the page loads
|
STRC_MOUSE
|
||||||
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STRC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.981568 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
STRC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
OTOAN_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 3) | NC score | 0.507624 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTW8, Q8NA86, Q96M76 | Gene names | OTOA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
OTOAN_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 4) | NC score | 0.494422 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
TARSH_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 5) | NC score | 0.086873 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 6) | NC score | 0.064309 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
MAGI2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.030899 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 8) | NC score | 0.028156 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.044285 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.061201 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.042087 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.006648 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
HPS5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.042409 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59438 | Gene names | Hps5, Ru2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein homolog (Ruby-eye protein 2) (Ru2). | |||||
|
PCDGD_HUMAN
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.009585 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
SEM6B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.015926 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.030634 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
FBX46_MOUSE
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.032825 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.032132 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.034366 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
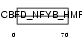
TAF6L_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.032409 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |
|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.041775 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
JPH1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.024189 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.029097 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.039714 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.038096 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.018423 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
LAP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.014257 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
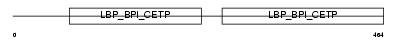
LPLC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.037435 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59827, Q5TDX6 | Gene names | LPLUNC4, C20orf186 | |||
|
Domain Architecture |
|
|||||
| Description | Long palate, lung and nasal epithelium carcinoma-associated protein 4 (Ligand-binding protein RY2G5). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.033858 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
PO6F1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.015094 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.031383 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.042473 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.020558 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
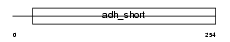
HCD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.023167 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.035360 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.015021 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.026201 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.029232 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.021014 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.015579 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.015560 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.015350 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.015302 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.029829 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.011797 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.028278 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.029647 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
TECT3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.021609 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2Q6, Q8CDF3, Q9CRC5 | Gene names | Tect3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-3 precursor. | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.025079 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.026312 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.052912 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CO026_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.025652 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P656, Q8N906 | Gene names | C15orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf26. | |||||
|
CP1A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.009036 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00184, Q61455 | Gene names | Cyp1a1, Cyp1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1). | |||||
|
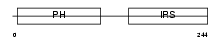
DOK2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.018303 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |
|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.025321 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
LAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.011891 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.014093 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
DYH5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.018362 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
LARP4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.019879 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.018840 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.020811 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NSMA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.017410 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.017442 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.006807 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZN217_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.003530 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.024621 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
C1QT6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.009562 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXI9 | Gene names | C1QTNF6, CTRP6 | |||
|
Domain Architecture |
|
|||||
| Description | Complement C1q tumor necrosis factor-related protein 6 precursor. | |||||
|
FMN1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.034321 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
FMN1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.032820 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
FOXP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.008695 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
HCD2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.018463 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.007408 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
MAGD4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.009619 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
PR285_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.028143 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
RM49_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.020800 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13405 | Gene names | MRPL49, C11orf4, NOF1 | |||
|
Domain Architecture |
|
|||||
| Description | Mitochondrial 39S ribosomal protein L49 (L49mt) (MRP-L49) (Protein NOF1) (Neighbor of FAU) (NOF). | |||||
|
SOX6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.009261 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
ZF276_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.001436 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.003667 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ANR16_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.003092 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.027528 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
G137B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.019306 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.006752 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
KCNC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.007195 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
MNT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.036916 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.002265 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.035616 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.012239 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.039144 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ZW10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.012089 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54692, Q3TIA5, Q3ULW1, Q921H3 | Gene names | Zw10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050797 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
STRC_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 89 | |
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
STRC_HUMAN
|
||||||
| NC score | 0.981568 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
OTOAN_HUMAN
|
||||||
| NC score | 0.507624 (rank : 3) | θ value | 3.51386e-20 (rank : 3) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTW8, Q8NA86, Q96M76 | Gene names | OTOA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
OTOAN_MOUSE
|
||||||
| NC score | 0.494422 (rank : 4) | θ value | 3.28887e-18 (rank : 4) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.086873 (rank : 5) | θ value | 0.00665767 (rank : 5) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.064309 (rank : 6) | θ value | 0.0330416 (rank : 6) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.061201 (rank : 7) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.052912 (rank : 8) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.050797 (rank : 9) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.044285 (rank : 10) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.042473 (rank : 11) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
HPS5_MOUSE
|
||||||
| NC score | 0.042409 (rank : 12) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59438 | Gene names | Hps5, Ru2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 5 protein homolog (Ruby-eye protein 2) (Ru2). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.042087 (rank : 13) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.041775 (rank : 14) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.039714 (rank : 15) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.039144 (rank : 16) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.038096 (rank : 17) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
LPLC4_HUMAN
|
||||||
| NC score | 0.037435 (rank : 18) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59827, Q5TDX6 | Gene names | LPLUNC4, C20orf186 | |||
|
Domain Architecture |

|
|||||
| Description | Long palate, lung and nasal epithelium carcinoma-associated protein 4 (Ligand-binding protein RY2G5). | |||||
|
MNT_HUMAN
|
||||||
| NC score | 0.036916 (rank : 19) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 496 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99583 | Gene names | MNT, ROX | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.035616 (rank : 20) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.035360 (rank : 21) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.034366 (rank : 22) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
FMN1A_MOUSE
|
||||||
| NC score | 0.034321 (rank : 23) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q05860 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoforms I/II/III (Limb deformity protein). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.033858 (rank : 24) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
FBX46_MOUSE
|
||||||
| NC score | 0.032825 (rank : 25) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BG80 | Gene names | Fbxo46, Fbx46, Fbxo34l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 46 (F-box only protein 34-like). | |||||
|
FMN1B_MOUSE
|
||||||
| NC score | 0.032820 (rank : 26) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q05859 | Gene names | Fmn, Ld | |||
|
Domain Architecture |

|
|||||
| Description | Formin-1 isoform IV (Limb deformity protein). | |||||
|
TAF6L_MOUSE
|
||||||
| NC score | 0.032409 (rank : 27) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R2K4, Q8C5T2 | Gene names | Taf6l, Paf65a | |||
|
Domain Architecture |

|
|||||
| Description | TAF6-like RNA polymerase II p300/CBP-associated factor-associated factor 65 kDa subunit 6L (PCAF-associated factor 65 alpha) (PAF65- alpha). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.032132 (rank : 28) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.031383 (rank : 29) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
MAGI2_MOUSE
|
||||||
| NC score | 0.030899 (rank : 30) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WVQ1, Q3UH81, Q6GT88, Q8BYT1, Q8CA85 | Gene names | Magi2, Acvrinp1, Aip1, Arip1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2 (Membrane-associated guanylate kinase inverted 2) (MAGI-2) (Atrophin-1-interacting protein 1) (Activin receptor-interacting protein 1) (Acvrip1). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.030634 (rank : 31) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.029829 (rank : 32) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.029647 (rank : 33) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.029232 (rank : 34) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.029097 (rank : 35) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.028278 (rank : 36) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.028156 (rank : 37) | θ value | 0.0961366 (rank : 8) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.028143 (rank : 38) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.027528 (rank : 39) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.026312 (rank : 40) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.026201 (rank : 41) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CO026_HUMAN
|
||||||
| NC score | 0.025652 (rank : 42) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P656, Q8N906 | Gene names | C15orf26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf26. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.025321 (rank : 43) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.025079 (rank : 44) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.024621 (rank : 45) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
JPH1_MOUSE
|
||||||
| NC score | 0.024189 (rank : 46) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9ET80, Q9EQZ3 | Gene names | Jph1, Jp1 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-1 (Junctophilin type 1) (JP-1). | |||||
|
HCD2_HUMAN
|
||||||
| NC score | 0.023167 (rank : 47) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99714, Q8TCV9, Q96HD5 | Gene names | HADH2, ERAB, HSD17B10, SCHAD, XH98G2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein) (Short-chain type dehydrogenase/reductase XH98G2). | |||||
|
TECT3_MOUSE
|
||||||
| NC score | 0.021609 (rank : 48) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2Q6, Q8CDF3, Q9CRC5 | Gene names | Tect3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tectonic-3 precursor. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.021014 (rank : 49) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.020811 (rank : 50) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RM49_HUMAN
|
||||||
| NC score | 0.020800 (rank : 51) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13405 | Gene names | MRPL49, C11orf4, NOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial 39S ribosomal protein L49 (L49mt) (MRP-L49) (Protein NOF1) (Neighbor of FAU) (NOF). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.020558 (rank : 52) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
LARP4_MOUSE
|
||||||
| NC score | 0.019879 (rank : 53) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BWW4, Q3UR69 | Gene names | Larp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | La-related protein 4 (La ribonucleoprotein domain family member 4). | |||||
|
G137B_HUMAN
|
||||||
| NC score | 0.019306 (rank : 54) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.018840 (rank : 55) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
HCD2_MOUSE
|
||||||
| NC score | 0.018463 (rank : 56) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08756 | Gene names | Hadh2, Erab, Hsd17b10 | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyacyl-CoA dehydrogenase type-2 (EC 1.1.1.35) (3-hydroxyacyl- CoA dehydrogenase type II) (Type II HADH) (3-hydroxy-2-methylbutyryl- CoA dehydrogenase) (EC 1.1.1.178) (Endoplasmic reticulum-associated amyloid beta-peptide-binding protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.018423 (rank : 57) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
DYH5_MOUSE
|
||||||
| NC score | 0.018362 (rank : 58) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VHE6, Q8BWG1 | Gene names | Dnah5, Dnahc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5). | |||||
|
DOK2_MOUSE
|
||||||
| NC score | 0.018303 (rank : 59) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70469, O70272, Q99KL1 | Gene names | Dok2, Frip | |||
|
Domain Architecture |

|
|||||
| Description | Docking protein 2 (Downstream of tyrosine kinase 2) (p56(dok-2)) (Dok- related protein) (Dok-R) (IL-four receptor-interacting protein) (FRIP). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.017442 (rank : 60) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
NSMA2_MOUSE
|
||||||
| NC score | 0.017410 (rank : 61) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JJY3, Q3UTQ5 | Gene names | smpd3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sphingomyelin phosphodiesterase 3 (EC 3.1.4.12) (Neutral sphingomyelinase 2) (Neutral sphingomyelinase II) (nSMase2) (nSMase- 2). | |||||
|
SEM6B_HUMAN
|
||||||
| NC score | 0.015926 (rank : 62) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H3T3, Q9NRK9 | Gene names | SEMA6B, SEMAZ | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin Z) (Sema Z). | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.015579 (rank : 63) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.015560 (rank : 64) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.015350 (rank : 65) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| NC score | 0.015302 (rank : 66) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
PO6F1_MOUSE
|
||||||
| NC score | 0.015094 (rank : 67) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q07916, Q91XK3 | Gene names | Pou6f1, Emb | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 1 (Octamer-binding transcription factor EMB) (Transcription regulatory protein MCP-1). | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.015021 (rank : 68) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
LAP2_HUMAN
|
||||||
| NC score | 0.014257 (rank : 69) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96RT1, Q86W38, Q9NR18, Q9NW48, Q9ULJ5 | Gene names | ERBB2IP, ERBIN, KIAA1225, LAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.014093 (rank : 70) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.012239 (rank : 71) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
ZW10_MOUSE
|
||||||
| NC score | 0.012089 (rank : 72) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54692, Q3TIA5, Q3ULW1, Q921H3 | Gene names | Zw10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere/kinetochore protein zw10 homolog. | |||||
|
LAP2_MOUSE
|
||||||
| NC score | 0.011891 (rank : 73) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.011797 (rank : 74) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
MAGD4_HUMAN
|
||||||
| NC score | 0.009619 (rank : 75) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96JG8, Q8WXW4, Q9BQ84, Q9BYH3, Q9BYH4 | Gene names | MAGED4, KIAA1859, MAGEE1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D4 (MAGE-D4 antigen) (MAGE-E1 antigen). | |||||
|
PCDGD_HUMAN
|
||||||
| NC score | 0.009585 (rank : 76) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
C1QT6_HUMAN
|
||||||
| NC score | 0.009562 (rank : 77) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXI9 | Gene names | C1QTNF6, CTRP6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q tumor necrosis factor-related protein 6 precursor. | |||||
|
SOX6_MOUSE
|
||||||
| NC score | 0.009261 (rank : 78) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40645, Q62250, Q9QWS5 | Gene names | Sox6, Sox-6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-6 (SOX-LZ). | |||||
|
CP1A1_MOUSE
|
||||||
| NC score | 0.009036 (rank : 79) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00184, Q61455 | Gene names | Cyp1a1, Cyp1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1). | |||||
|
FOXP4_MOUSE
|
||||||
| NC score | 0.008695 (rank : 80) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9DBY0, Q80V92, Q8CG10, Q8CIS1 | Gene names | Foxp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein P4 (Fork head-related protein like A) (mFKHLA). | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.007408 (rank : 81) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
KCNC3_HUMAN
|
||||||
| NC score | 0.007195 (rank : 82) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14003 | Gene names | KCNC3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily C member 3 (Voltage-gated potassium channel subunit Kv3.3) (KSHIIID). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.006807 (rank : 83) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.006752 (rank : 84) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.006648 (rank : 85) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.003667 (rank : 86) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
ZN217_HUMAN
|
||||||
| NC score | 0.003530 (rank : 87) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75362 | Gene names | ZNF217, ZABC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 217. | |||||
|
ANR16_HUMAN
|
||||||
| NC score | 0.003092 (rank : 88) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.002265 (rank : 89) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ZF276_HUMAN
|
||||||
| NC score | 0.001436 (rank : 90) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 89 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N554 | Gene names | ZFP276 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 276 homolog (Zfp-276). | |||||