Please be patient as the page loads
|
COT1_MOUSE
|
||||||
| SwissProt Accessions | Q60632, Q61438 | Gene names | Nr2f1, Erbal3, Tfcoup1 | |||
|
Domain Architecture |
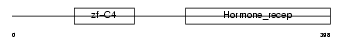
|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999389 (rank : 2) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 147 | |
| SwissProt Accessions | P10589 | Gene names | NR2F1, EAR3, ERBAL3, TFCOUP1 | |||
|
Domain Architecture |
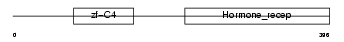
|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I) (V-ERBA-related protein EAR-3). | |||||
|
COT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 160 | |
| SwissProt Accessions | Q60632, Q61438 | Gene names | Nr2f1, Erbal3, Tfcoup1 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I). | |||||
|
COT2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998128 (rank : 3) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P24468, Q03754 | Gene names | NR2F2, ARP1, TFCOUP2 | |||
|
Domain Architecture |
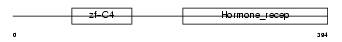
|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
COT2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998128 (rank : 4) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P43135 | Gene names | Nr2f2, Aporp1, Arp-1, Arp1, Tfcoup2 | |||
|
Domain Architecture |
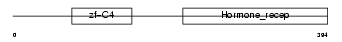
|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
EAR2_HUMAN
|
||||||
| θ value | 2.87295e-139 (rank : 5) | NC score | 0.995521 (rank : 5) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P10588, Q9BUE8 | Gene names | NR2F6, EAR2, ERBAL2 | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor EAR-2 (V-erbA-related protein EAR-2). | |||||
|
EAR2_MOUSE
|
||||||
| θ value | 7.07634e-138 (rank : 6) | NC score | 0.995434 (rank : 6) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P43136, Q61504 | Gene names | Nr2f6, Ear-2, Ear2, Erbal2 | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor EAR-2 (V-erbA-related protein EAR-2). | |||||
|
RXRA_HUMAN
|
||||||
| θ value | 1.45253e-74 (rank : 7) | NC score | 0.975961 (rank : 14) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
RXRA_MOUSE
|
||||||
| θ value | 1.45253e-74 (rank : 8) | NC score | 0.978885 (rank : 13) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P28700 | Gene names | Rxra, Nr2b1 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
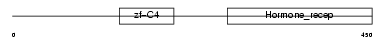
RXRG_HUMAN
|
||||||
| θ value | 5.51964e-74 (rank : 9) | NC score | 0.979875 (rank : 11) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | P48443 | Gene names | RXRG, NR2B3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
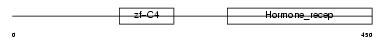
RXRG_MOUSE
|
||||||
| θ value | 1.04096e-72 (rank : 10) | NC score | 0.979652 (rank : 12) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P28705 | Gene names | Rxrg, Nr2b3 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
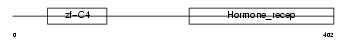
NR2E3_HUMAN
|
||||||
| θ value | 3.95564e-72 (rank : 11) | NC score | 0.984481 (rank : 7) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9Y5X4, Q9UHM4 | Gene names | NR2E3, PNR, RNR | |||
|
Domain Architecture |
|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
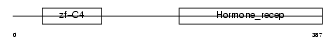
NR2E3_MOUSE
|
||||||
| θ value | 1.66191e-70 (rank : 12) | NC score | 0.983777 (rank : 8) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |
|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
RXRB_HUMAN
|
||||||
| θ value | 2.3998e-69 (rank : 13) | NC score | 0.971308 (rank : 20) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | P28702, P28703 | Gene names | RXRB, NR2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta). | |||||
|
RXRB_MOUSE
|
||||||
| θ value | 5.34618e-69 (rank : 14) | NC score | 0.974917 (rank : 16) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
NR2E1_HUMAN
|
||||||
| θ value | 1.19101e-68 (rank : 15) | NC score | 0.982744 (rank : 9) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9Y466 | Gene names | NR2E1, TLX | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (hTll). | |||||
|
NR2E1_MOUSE
|
||||||
| θ value | 1.5555e-68 (rank : 16) | NC score | 0.982668 (rank : 10) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q64104 | Gene names | Nr2e1, Tll, Tlx | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (mTll). | |||||
|
HNF4A_MOUSE
|
||||||
| θ value | 2.48917e-58 (rank : 17) | NC score | 0.974763 (rank : 18) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P49698 | Gene names | Hnf4a, Hnf-4, Hnf4, Nr2a1, Tcf14 | |||
|
Domain Architecture |
|
|||||
| Description | Hepatocyte nuclear factor 4-alpha (HNF-4-alpha) (Transcription factor HNF-4) (Transcription factor 14). | |||||
|
HNF4A_HUMAN
|
||||||
| θ value | 4.69435e-57 (rank : 18) | NC score | 0.974271 (rank : 19) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P41235, O00659, O00723, Q14540, Q92653, Q92654, Q92655, Q99864, Q9NQH0 | Gene names | HNF4A, HNF4, NR2A1, TCF14 | |||
|
Domain Architecture |
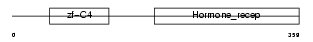
|
|||||
| Description | Hepatocyte nuclear factor 4-alpha (HNF-4-alpha) (Transcription factor HNF-4) (Transcription factor 14). | |||||
|
HNF4G_MOUSE
|
||||||
| θ value | 1.78386e-56 (rank : 19) | NC score | 0.974804 (rank : 17) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9WUU6 | Gene names | Hnf4g, Nr2a2 | |||
|
Domain Architecture |
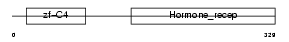
|
|||||
| Description | Hepatocyte nuclear factor 4-gamma (HNF-4-gamma). | |||||
|
HNF4G_HUMAN
|
||||||
| θ value | 3.0428e-56 (rank : 20) | NC score | 0.974974 (rank : 15) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q14541, Q9UH81, Q9UIS6 | Gene names | HNF4G, NR2A2 | |||
|
Domain Architecture |
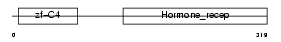
|
|||||
| Description | Hepatocyte nuclear factor 4-gamma (HNF-4-gamma). | |||||
|
ERR2_MOUSE
|
||||||
| θ value | 1.04822e-48 (rank : 21) | NC score | 0.955353 (rank : 21) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q61539, O09067, O09146 | Gene names | Esrrb, Err-2, Err2, Nr3b2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2). | |||||
|
ERR2_HUMAN
|
||||||
| θ value | 1.36902e-48 (rank : 22) | NC score | 0.952977 (rank : 24) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O95718, Q9HCB4 | Gene names | ESRRB, ERRB2, NR3B2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2) (ERR beta-2). | |||||
|
ERR3_HUMAN
|
||||||
| θ value | 6.7944e-48 (rank : 23) | NC score | 0.955132 (rank : 22) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P62508, O75454, O96021, Q68DA0, Q6P274, Q6PK28, Q6TS38, Q9R1F3, Q9UNJ4 | Gene names | ESRRG, ERR3, ERRG2, KIAA0832, NR3B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3) (ERR gamma-2). | |||||
|
ERR3_MOUSE
|
||||||
| θ value | 6.7944e-48 (rank : 24) | NC score | 0.955132 (rank : 23) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P62509, O75454, O96021, Q8CHC9, Q9R1F3 | Gene names | Esrrg, Err3, Kiaa0832, Nr3b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3). | |||||
|
ERR1_HUMAN
|
||||||
| θ value | 1.56636e-44 (rank : 25) | NC score | 0.952375 (rank : 25) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P11474, Q14514 | Gene names | ESRRA, ERR1, ESRL1, NR3B1 | |||
|
Domain Architecture |
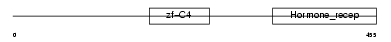
|
|||||
| Description | Steroid hormone receptor ERR1 (Estrogen-related receptor, alpha) (ERR- alpha) (Estrogen receptor-like 1). | |||||
|
NR4A2_HUMAN
|
||||||
| θ value | 5.57106e-42 (rank : 26) | NC score | 0.938994 (rank : 27) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P43354, Q16311 | Gene names | NR4A2, NOT, NURR1, TINUR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (Immediate-early response protein NOT) (Transcriptionally-inducible nuclear receptor). | |||||
|
NR4A2_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 27) | NC score | 0.939035 (rank : 26) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q06219, O08690 | Gene names | Nr4a2, Nurr1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (NUR- related factor 1). | |||||
|
NR4A1_HUMAN
|
||||||
| θ value | 1.79215e-40 (rank : 28) | NC score | 0.938676 (rank : 28) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P22736 | Gene names | NR4A1, GFRP1, HMR, NAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Early response protein NAK1) (TR3 orphan receptor) (ST-59). | |||||
|
RARG1_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 29) | NC score | 0.933020 (rank : 37) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
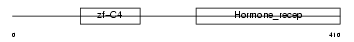
|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARG2_HUMAN
|
||||||
| θ value | 2.34062e-40 (rank : 30) | NC score | 0.933165 (rank : 35) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
NR4A1_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 31) | NC score | 0.937378 (rank : 29) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P12813 | Gene names | Nr4a1, Gfrp, Hmr, N10, Nur77 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Nuclear hormone receptor NUR/77) (N10 nuclear protein). | |||||
|
RARB_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 32) | NC score | 0.932591 (rank : 40) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P22605, P11417, P22604 | Gene names | Rarb, Nr1b2 | |||
|
Domain Architecture |
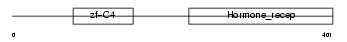
|
|||||
| Description | Retinoic acid receptor beta (RAR-beta). | |||||
|
RARG1_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 33) | NC score | 0.932336 (rank : 41) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
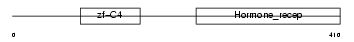
|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
RARG2_MOUSE
|
||||||
| θ value | 1.98146e-39 (rank : 34) | NC score | 0.932626 (rank : 39) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |
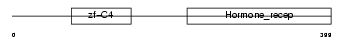
|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RARA_HUMAN
|
||||||
| θ value | 4.41421e-39 (rank : 35) | NC score | 0.933179 (rank : 34) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARA_MOUSE
|
||||||
| θ value | 7.52953e-39 (rank : 36) | NC score | 0.933084 (rank : 36) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P11416, P22603 | Gene names | Rara, Nr1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARB_HUMAN
|
||||||
| θ value | 7.52953e-39 (rank : 37) | NC score | 0.932631 (rank : 38) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P10826, P12891, Q00989, Q15298, Q9UN48 | Gene names | RARB, HAP, NR1B2 | |||
|
Domain Architecture |
|
|||||
| Description | Retinoic acid receptor beta (RAR-beta) (RAR-epsilon) (HBV-activated protein). | |||||
|
NR4A3_MOUSE
|
||||||
| θ value | 2.86122e-38 (rank : 38) | NC score | 0.936514 (rank : 30) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9QZB6, Q9QZB5 | Gene names | Nr4a3, Tec | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Orphan nuclear receptor TEC) (Translocated in extraskeletal chondrosarcoma). | |||||
|
NR4A3_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 39) | NC score | 0.935238 (rank : 31) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q92570, Q12935, Q14979, Q16420, Q4VXA9, Q9UEK2, Q9UEK3 | Gene names | NR4A3, CHN, CSMF, MINOR, NOR1, TEC | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Nuclear hormone receptor NOR-1) (Neuron-derived orphan receptor 1) (Mitogen-induced nuclear orphan receptor). | |||||
|
TR4_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 40) | NC score | 0.933955 (rank : 32) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P49116, P55092 | Gene names | NR2C2, TAK1, TR4 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor TR4 (Orphan nuclear receptor TAK1). | |||||
|
TR4_MOUSE
|
||||||
| θ value | 1.32909e-35 (rank : 41) | NC score | 0.933597 (rank : 33) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P49117, P55093 | Gene names | Nr2c2, Mtr2r1, Tak1, Tr4 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor TR4 (Orphan nuclear receptor TAK1). | |||||
|
ESR1_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 42) | NC score | 0.928482 (rank : 47) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
ESR1_MOUSE
|
||||||
| θ value | 2.34606e-32 (rank : 43) | NC score | 0.928561 (rank : 46) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P19785, Q9JJT5, Q9QY51, Q9QY52 | Gene names | Esr1, Esr, Estr, Estra, Nr3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
NR1H3_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 44) | NC score | 0.923489 (rank : 50) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q13133, Q96H87 | Gene names | NR1H3, LXRA | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-alpha (Liver X receptor alpha) (Nuclear orphan receptor LXR-alpha). | |||||
|
NR1H3_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 45) | NC score | 0.922248 (rank : 51) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Z0Y9, Q9QUH7 | Gene names | Nr1h3, Lxra | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-alpha (Liver X receptor alpha) (Nuclear orphan receptor LXR-alpha). | |||||
|
TR2_MOUSE
|
||||||
| θ value | 1.16434e-31 (rank : 46) | NC score | 0.930973 (rank : 43) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q505F1, P97763, Q3UIJ7, Q4U1Z4, Q60927, Q62152, Q8VIJ3 | Gene names | Nr2c1, Tr2, Tr2-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Orphan nuclear receptor TR2 (Testicular receptor 2) (mTR2). | |||||
|
PPARD_MOUSE
|
||||||
| θ value | 5.77852e-31 (rank : 47) | NC score | 0.913146 (rank : 63) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P35396, P37239 | Gene names | Ppard, Nr1c2, Pparb | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor delta (PPAR-delta) (PPAR- beta) (Nuclear hormone receptor 1) (NUC1). | |||||
|
PPARA_HUMAN
|
||||||
| θ value | 9.8567e-31 (rank : 48) | NC score | 0.915841 (rank : 58) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q07869, Q16241, Q92486, Q92689, Q9Y3N1 | Gene names | PPARA, NR1C1, PPAR | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor alpha (PPAR-alpha). | |||||
|
PPARD_HUMAN
|
||||||
| θ value | 1.28732e-30 (rank : 49) | NC score | 0.914488 (rank : 61) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q03181, Q7Z5K0, Q9BUD4 | Gene names | PPARD, NR1C2, PPARB | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor delta (PPAR-delta) (PPAR- beta) (Nuclear hormone receptor 1) (NUC1) (NUCI). | |||||
|
ESR2_MOUSE
|
||||||
| θ value | 1.6813e-30 (rank : 50) | NC score | 0.930822 (rank : 44) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O08537, O35635, O70519 | Gene names | Esr2, Estrb, Nr3a2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
PPARG_MOUSE
|
||||||
| θ value | 2.19584e-30 (rank : 51) | NC score | 0.911535 (rank : 67) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P37238 | Gene names | Pparg, Nr1c3 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma (PPAR-gamma). | |||||
|
PPARG_HUMAN
|
||||||
| θ value | 2.86786e-30 (rank : 52) | NC score | 0.911906 (rank : 65) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P37231, O00684, O00710, O14515, Q15178, Q15179, Q15180, Q15832, Q86U60, Q96J12 | Gene names | PPARG, NR1C3 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma (PPAR-gamma). | |||||
|
ESR2_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 53) | NC score | 0.932195 (rank : 42) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q92731, O60608, O60685, O60702, O60703, O75583, O75584, Q86Z31, Q9UEV6, Q9UHD3, Q9UQK9 | Gene names | ESR2, ESTRB, NR3A2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
PPARA_MOUSE
|
||||||
| θ value | 4.89182e-30 (rank : 54) | NC score | 0.914873 (rank : 60) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P23204 | Gene names | Ppara, Nr1c1, Ppar | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor alpha (PPAR-alpha). | |||||
|
NR1H2_MOUSE
|
||||||
| θ value | 1.08979e-29 (rank : 55) | NC score | 0.913820 (rank : 62) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
NR1H4_MOUSE
|
||||||
| θ value | 5.40856e-29 (rank : 56) | NC score | 0.920658 (rank : 55) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q60641, Q60642, Q60643 | Gene names | Nr1h4, Bar, Fxr, Rip14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
TR2_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 57) | NC score | 0.928689 (rank : 45) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P13056 | Gene names | NR2C1, TR2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor TR2 (Testicular receptor 2). | |||||
|
NR5A2_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 58) | NC score | 0.921678 (rank : 53) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P45448 | Gene names | Nr5a2, Lrh1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR5A2 (Liver receptor homolog 1) (LRH-1). | |||||
|
NR1H4_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 59) | NC score | 0.921368 (rank : 54) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q96RI1, Q8NFP5, Q8NFP6, Q92943 | Gene names | NR1H4, BAR, FXR, HRR1, RIP14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
THA_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 60) | NC score | 0.909449 (rank : 68) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P10827, P21205, Q8N6A1, Q96H73 | Gene names | THRA, ERBA1, NR1A1, THRA1 | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid hormone receptor alpha (C-erbA-alpha) (c-erbA-1) (EAR-7) (EAR7). | |||||
|
THA_MOUSE
|
||||||
| θ value | 1.33217e-27 (rank : 61) | NC score | 0.909443 (rank : 69) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P63058, P10685, P15827, P16416, P37241, Q63107, Q63195, Q63196, Q99146 | Gene names | Thra, C-erba-alpha, Nr1a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor alpha (C-erbA-alpha) (c-erbA-1). | |||||
|
NR1I3_HUMAN
|
||||||
| θ value | 1.12775e-26 (rank : 62) | NC score | 0.922187 (rank : 52) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q14994 | Gene names | NR1I3, CAR | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor NR1I3 (Constitutive androstane receptor) (CAR) (Constitutive activator of retinoid response) (Constitutive active response) (Orphan nuclear receptor MB67). | |||||
|
NR5A2_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 63) | NC score | 0.918438 (rank : 56) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O00482, O95642 | Gene names | NR5A2, B1F, CPF, FTF | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR5A2 (Alpha-1-fetoprotein transcription factor) (Hepatocytic transcription factor) (B1-binding factor) (hB1F) (CYP7A promoter-binding factor) (Liver receptor homolog 1) (LRH-1). | |||||
|
RORG_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 64) | NC score | 0.898792 (rank : 80) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P51449, Q8N5V7, Q8NCY8 | Gene names | RORC, NR1F3, RORG, RZRG | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-gamma (Nuclear receptor RZR-gamma). | |||||
|
RORG_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 65) | NC score | 0.902022 (rank : 75) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P51450, Q61027, Q91YT5, Q9QXD9, Q9R177 | Gene names | Rorc, Nr1f3, Rorg, Thor | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-gamma (Nuclear receptor RZR-gamma) (Thymus orphan receptor) (TOR). | |||||
|
RORA_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 66) | NC score | 0.897906 (rank : 81) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P35398, P35397, P35399, P45445, Q96H83 | Gene names | RORA, NR1F1, RZRA | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-alpha (Nuclear receptor RZR-alpha). | |||||
|
RORA_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 67) | NC score | 0.897877 (rank : 82) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P51448, P70283, P97741, P97773, Q923G1 | Gene names | Rora, Nr1f1, Rzra | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-alpha (Nuclear receptor RZR-alpha). | |||||
|
RORB_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 68) | NC score | 0.899256 (rank : 78) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q92753, Q8WX73 | Gene names | RORB, NR1F2, RZRB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-beta (Nuclear receptor RZR-beta). | |||||
|
RORB_MOUSE
|
||||||
| θ value | 4.73814e-25 (rank : 69) | NC score | 0.899028 (rank : 79) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8R1B8 | Gene names | Rorb, Nr1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-beta (Nuclear receptor RZR-beta). | |||||
|
THB1_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 70) | NC score | 0.903274 (rank : 73) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P10828, Q13986 | Gene names | THRB, ERBA2, NR1A2, THR1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-1. | |||||
|
THB2_HUMAN
|
||||||
| θ value | 8.08199e-25 (rank : 71) | NC score | 0.903702 (rank : 72) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P37243 | Gene names | THRB, ERBA2, NR1A2, THR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor beta-2. | |||||
|
NR1I3_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 72) | NC score | 0.917588 (rank : 57) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O35627, O35628 | Gene names | Nr1i3, Car | |||
|
Domain Architecture |
|
|||||
| Description | Orphan nuclear receptor NR1I3 (Constitutive androstane receptor) (CAR) (Orphan nuclear receptor MB67). | |||||
|
THB1_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 73) | NC score | 0.901910 (rank : 76) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P37242 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-1. | |||||
|
THB2_MOUSE
|
||||||
| θ value | 5.23862e-24 (rank : 74) | NC score | 0.901774 (rank : 77) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P37244 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-2. | |||||
|
ANDR_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 75) | NC score | 0.876678 (rank : 89) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P10275 | Gene names | AR, DHTR, NR3C4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
GCR_HUMAN
|
||||||
| θ value | 3.39556e-23 (rank : 76) | NC score | 0.880433 (rank : 87) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P04150, P04151, Q53EP5, Q6N0A4 | Gene names | NR3C1, GRL | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor (GR). | |||||
|
MCR_HUMAN
|
||||||
| θ value | 4.43474e-23 (rank : 77) | NC score | 0.871854 (rank : 91) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P08235, Q96KQ8, Q96KQ9 | Gene names | NR3C2, MCR, MLR | |||
|
Domain Architecture |

|
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
ANDR_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 78) | NC score | 0.877868 (rank : 88) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
NR1D2_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 79) | NC score | 0.902566 (rank : 74) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q60674, Q60646 | Gene names | Nr1d2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (Orphan nuclear receptor RVR). | |||||
|
NR6A1_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 80) | NC score | 0.925559 (rank : 49) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q64249 | Gene names | Nr6a1, Gcnf | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR6A1 (Germ cell nuclear factor) (GCNF) (Retinoid receptor-related testis-specific receptor) (RTR). | |||||
|
MCR_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 81) | NC score | 0.868120 (rank : 93) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
NR1D2_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 82) | NC score | 0.904800 (rank : 71) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q14995, O00402, Q86XD4 | Gene names | NR1D2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (EAR-1R) (Orphan nuclear hormone receptor BD73). | |||||
|
NR1D1_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 83) | NC score | 0.911609 (rank : 66) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
STF1_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 84) | NC score | 0.912876 (rank : 64) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q13285, O15196 | Gene names | NR5A1, AD4BP, FTZF1, SF1 | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic factor 1 (STF-1) (SF-1) (Adrenal 4-binding protein) (Steroid hormone receptor Ad4BP) (Fushi tarazu factor homolog 1). | |||||
|
STF1_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 85) | NC score | 0.915578 (rank : 59) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P33242 | Gene names | Nr5a1, Ftzf1 | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic factor 1 (STF-1) (SF-1) (Adrenal 4-binding protein) (Steroid hormone receptor Ad4BP) (Fushi tarazu factor homolog 1) (Embryonal LTR-binding protein) (ELP) (Embryonal long terminal repeat- binding protein) (Steroid hydroxylase positive regulator). | |||||
|
NR1H2_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 86) | NC score | 0.905507 (rank : 70) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P55055, Q12970 | Gene names | NR1H2, LXRB, NER, UNR | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Nuclear receptor NER). | |||||
|
NR6A1_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 87) | NC score | 0.926638 (rank : 48) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q15406, O00551, O00603, Q8NHQ0, Q92898, Q99802 | Gene names | NR6A1, GCNF | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR6A1 (Germ cell nuclear factor) (GCNF) (Retinoid receptor-related testis-specific receptor) (RTR). | |||||
|
PRGR_HUMAN
|
||||||
| θ value | 1.86321e-21 (rank : 88) | NC score | 0.869424 (rank : 92) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
PRGR_MOUSE
|
||||||
| θ value | 3.17815e-21 (rank : 89) | NC score | 0.864218 (rank : 94) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
SHP_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 90) | NC score | 0.646185 (rank : 95) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15466, Q5QP36 | Gene names | NR0B2, SHP | |||
|
Domain Architecture |
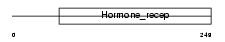
|
|||||
| Description | Nuclear receptor 0B2 (Orphan nuclear receptor SHP) (Small heterodimer partner). | |||||
|
GCR_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 91) | NC score | 0.873392 (rank : 90) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P06537, Q61628, Q61629 | Gene names | Nr3c1, Grl, Grl1 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor (GR). | |||||
|
SHP_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 92) | NC score | 0.639195 (rank : 96) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62227, Q53Z53 | Gene names | Nr0b2, Shp | |||
|
Domain Architecture |
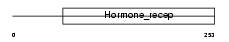
|
|||||
| Description | Nuclear receptor 0B2 (Orphan nuclear receptor SHP) (Small heterodimer partner). | |||||
|
VDR_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 93) | NC score | 0.895799 (rank : 84) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P11473 | Gene names | VDR, NR1I1 | |||
|
Domain Architecture |
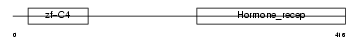
|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
VDR_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 94) | NC score | 0.897309 (rank : 83) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P48281 | Gene names | Vdr, Nr1i1 | |||
|
Domain Architecture |
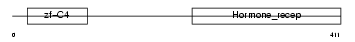
|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
PXR_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 95) | NC score | 0.892596 (rank : 86) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O75469, Q96AC7, Q9UJ22, Q9UJ23, Q9UJ24, Q9UJ25, Q9UJ26, Q9UJ27 | Gene names | NR1I2, PXR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor PXR (Pregnane X receptor) (Orphan nuclear receptor PAR1) (Steroid and xenobiotic receptor) (SXR). | |||||
|
PXR_MOUSE
|
||||||
| θ value | 3.07829e-16 (rank : 96) | NC score | 0.894664 (rank : 85) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O54915 | Gene names | Nr1i2, Pxr | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor PXR (Pregnane X receptor). | |||||
|
DAX1_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 97) | NC score | 0.592779 (rank : 98) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51843, Q96F69 | Gene names | NR0B1, AHC, DAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor 0B1 (Nuclear receptor DAX-1) (DSS-AHC critical region on the X chromosome protein 1). | |||||
|
DAX1_MOUSE
|
||||||
| θ value | 6.00763e-12 (rank : 98) | NC score | 0.610337 (rank : 97) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61066, O09068, O09147 | Gene names | Nr0b1, Ahch, Dax1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor 0B1 (Nuclear receptor DAX-1). | |||||
|
CO4A2_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 99) | NC score | 0.045550 (rank : 101) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 100) | NC score | 0.045797 (rank : 100) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 101) | NC score | 0.044376 (rank : 105) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO7A1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 102) | NC score | 0.029203 (rank : 129) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 103) | NC score | 0.045117 (rank : 103) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
C1QC_MOUSE
|
||||||
| θ value | 0.125558 (rank : 104) | NC score | 0.035642 (rank : 121) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02105 | Gene names | C1qc, C1qg | |||
|
Domain Architecture |
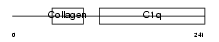
|
|||||
| Description | Complement C1q subcomponent subunit C precursor. | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 105) | NC score | 0.040676 (rank : 111) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 106) | NC score | 0.052274 (rank : 99) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 107) | NC score | 0.044432 (rank : 104) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 108) | NC score | 0.040536 (rank : 112) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 109) | NC score | 0.039016 (rank : 113) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 110) | NC score | 0.045328 (rank : 102) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CO2A1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 111) | NC score | 0.043200 (rank : 106) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |
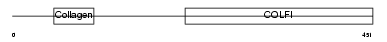
|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO2A1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 112) | NC score | 0.042605 (rank : 108) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO4A4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 113) | NC score | 0.042249 (rank : 109) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |
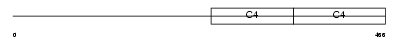
|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
CEL_HUMAN
|
||||||
| θ value | 1.06291 (rank : 114) | NC score | 0.015545 (rank : 135) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 115) | NC score | -0.007345 (rank : 158) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
CO6A1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 116) | NC score | 0.033083 (rank : 126) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |
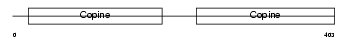
|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 1.38821 (rank : 117) | NC score | -0.006372 (rank : 157) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 118) | NC score | 0.042718 (rank : 107) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
BOC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 119) | NC score | -0.000786 (rank : 154) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 120) | NC score | 0.038333 (rank : 115) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 121) | NC score | 0.041657 (rank : 110) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 122) | NC score | -0.002787 (rank : 155) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 123) | NC score | 0.006677 (rank : 142) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 124) | NC score | 0.033535 (rank : 125) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
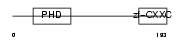
CXCC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 125) | NC score | 0.006553 (rank : 143) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |
|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 126) | NC score | 0.004576 (rank : 146) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
RTEL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 127) | NC score | 0.005490 (rank : 144) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
CO1A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 128) | NC score | 0.038097 (rank : 116) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 129) | NC score | 0.037163 (rank : 119) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
COAA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 130) | NC score | 0.034564 (rank : 123) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q05306 | Gene names | Col10a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
COEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 131) | NC score | 0.022107 (rank : 133) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
CONA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 132) | NC score | 0.034671 (rank : 122) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CONA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 133) | NC score | 0.037743 (rank : 118) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K4G2, Q5SUQ0 | Gene names | Col23a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
KCNH6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 134) | NC score | 0.001753 (rank : 150) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 135) | NC score | 0.002046 (rank : 149) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
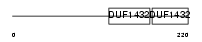
PODO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 136) | NC score | 0.007063 (rank : 141) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NP85, Q8N6Q5 | Gene names | NPHS2 | |||
|
Domain Architecture |
|
|||||
| Description | Podocin. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 4.03905 (rank : 137) | NC score | 0.019901 (rank : 134) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CO4A6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 138) | NC score | 0.038984 (rank : 114) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
SI1L1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 139) | NC score | 0.004868 (rank : 145) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
ZN341_HUMAN
|
||||||
| θ value | 5.27518 (rank : 140) | NC score | -0.005559 (rank : 156) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYN7, Q5JXM8, Q96ST5 | Gene names | ZNF341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 341. | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 6.88961 (rank : 141) | NC score | 0.032413 (rank : 128) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
C1QT5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 142) | NC score | 0.029093 (rank : 130) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K479 | Gene names | C1qtnf5, Ctrp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1q tumor necrosis factor-related protein 5 precursor. | |||||
|
CO6A1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 143) | NC score | 0.032565 (rank : 127) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q04857 | Gene names | Col6a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 144) | NC score | 0.035815 (rank : 120) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 145) | NC score | 0.033610 (rank : 124) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
ERB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 146) | NC score | 0.008220 (rank : 140) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 147) | NC score | -0.008597 (rank : 160) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
RMD5B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 148) | NC score | 0.009709 (rank : 138) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YQ7 | Gene names | Rmnd5b | |||
|
Domain Architecture |

|
|||||
| Description | RMD5 homolog B. | |||||
|
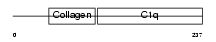
ADIPO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.023424 (rank : 131) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15848 | Gene names | ADIPOQ, ACDC, ACRP30, APM1, GBP28 | |||
|
Domain Architecture |
|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipose most abundant gene transcript 1) (apM-1) (Gelatin-binding protein). | |||||
|
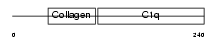
ADIPO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.023168 (rank : 132) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60994, Q62400, Q9DC68 | Gene names | Adipoq, Acdc, Acrp30, Apm1 | |||
|
Domain Architecture |
|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipocyte-specific protein AdipoQ). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.000600 (rank : 152) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.037892 (rank : 117) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CXCC1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.004285 (rank : 147) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
FA62A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.000996 (rank : 151) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FCN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.013301 (rank : 136) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70165 | Gene names | Fcn1, Fcna | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-1 precursor (Collagen/fibrinogen domain-containing protein 1) (Ficolin-A) (Ficolin A) (M-Ficolin). | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | -0.000563 (rank : 153) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.010330 (rank : 137) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PKN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | -0.008416 (rank : 159) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
RMD5B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.008723 (rank : 139) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96G75, Q6UVY7, Q9H6F6 | Gene names | RMND5B | |||
|
Domain Architecture |

|
|||||
| Description | RMD5 homolog B. | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.004153 (rank : 148) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
COT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 160 | |
| SwissProt Accessions | Q60632, Q61438 | Gene names | Nr2f1, Erbal3, Tfcoup1 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I). | |||||
|
COT1_HUMAN
|
||||||
| NC score | 0.999389 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 147 | |
| SwissProt Accessions | P10589 | Gene names | NR2F1, EAR3, ERBAL3, TFCOUP1 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 1 (COUP-TF1) (COUP-TF I) (V-ERBA-related protein EAR-3). | |||||
|
COT2_HUMAN
|
||||||
| NC score | 0.998128 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P24468, Q03754 | Gene names | NR2F2, ARP1, TFCOUP2 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
COT2_MOUSE
|
||||||
| NC score | 0.998128 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P43135 | Gene names | Nr2f2, Aporp1, Arp-1, Arp1, Tfcoup2 | |||
|
Domain Architecture |

|
|||||
| Description | COUP transcription factor 2 (COUP-TF2) (COUP-TF II) (Apolipoprotein AI regulatory protein 1) (ARP-1). | |||||
|
EAR2_HUMAN
|
||||||
| NC score | 0.995521 (rank : 5) | θ value | 2.87295e-139 (rank : 5) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P10588, Q9BUE8 | Gene names | NR2F6, EAR2, ERBAL2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor EAR-2 (V-erbA-related protein EAR-2). | |||||
|
EAR2_MOUSE
|
||||||
| NC score | 0.995434 (rank : 6) | θ value | 7.07634e-138 (rank : 6) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P43136, Q61504 | Gene names | Nr2f6, Ear-2, Ear2, Erbal2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor EAR-2 (V-erbA-related protein EAR-2). | |||||
|
NR2E3_HUMAN
|
||||||
| NC score | 0.984481 (rank : 7) | θ value | 3.95564e-72 (rank : 11) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9Y5X4, Q9UHM4 | Gene names | NR2E3, PNR, RNR | |||
|
Domain Architecture |

|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
NR2E3_MOUSE
|
||||||
| NC score | 0.983777 (rank : 8) | θ value | 1.66191e-70 (rank : 12) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |

|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
NR2E1_HUMAN
|
||||||
| NC score | 0.982744 (rank : 9) | θ value | 1.19101e-68 (rank : 15) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9Y466 | Gene names | NR2E1, TLX | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (hTll). | |||||
|
NR2E1_MOUSE
|
||||||
| NC score | 0.982668 (rank : 10) | θ value | 1.5555e-68 (rank : 16) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q64104 | Gene names | Nr2e1, Tll, Tlx | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR2E1 (Nuclear receptor TLX) (Tailless homolog) (Tll) (mTll). | |||||
|
RXRG_HUMAN
|
||||||
| NC score | 0.979875 (rank : 11) | θ value | 5.51964e-74 (rank : 9) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | P48443 | Gene names | RXRG, NR2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
RXRG_MOUSE
|
||||||
| NC score | 0.979652 (rank : 12) | θ value | 1.04096e-72 (rank : 10) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P28705 | Gene names | Rxrg, Nr2b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
RXRA_MOUSE
|
||||||
| NC score | 0.978885 (rank : 13) | θ value | 1.45253e-74 (rank : 8) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | P28700 | Gene names | Rxra, Nr2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
RXRA_HUMAN
|
||||||
| NC score | 0.975961 (rank : 14) | θ value | 1.45253e-74 (rank : 7) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P19793, Q2V504 | Gene names | RXRA, NR2B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-alpha (Retinoid X receptor alpha). | |||||
|
HNF4G_HUMAN
|
||||||
| NC score | 0.974974 (rank : 15) | θ value | 3.0428e-56 (rank : 20) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q14541, Q9UH81, Q9UIS6 | Gene names | HNF4G, NR2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 4-gamma (HNF-4-gamma). | |||||
|
RXRB_MOUSE
|
||||||
| NC score | 0.974917 (rank : 16) | θ value | 5.34618e-69 (rank : 14) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P28704, P33243 | Gene names | Rxrb, Nr2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta) (MHC class I regulatory element-binding protein H-2RIIBP). | |||||
|
HNF4G_MOUSE
|
||||||
| NC score | 0.974804 (rank : 17) | θ value | 1.78386e-56 (rank : 19) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q9WUU6 | Gene names | Hnf4g, Nr2a2 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 4-gamma (HNF-4-gamma). | |||||
|
HNF4A_MOUSE
|
||||||
| NC score | 0.974763 (rank : 18) | θ value | 2.48917e-58 (rank : 17) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P49698 | Gene names | Hnf4a, Hnf-4, Hnf4, Nr2a1, Tcf14 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 4-alpha (HNF-4-alpha) (Transcription factor HNF-4) (Transcription factor 14). | |||||
|
HNF4A_HUMAN
|
||||||
| NC score | 0.974271 (rank : 19) | θ value | 4.69435e-57 (rank : 18) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P41235, O00659, O00723, Q14540, Q92653, Q92654, Q92655, Q99864, Q9NQH0 | Gene names | HNF4A, HNF4, NR2A1, TCF14 | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 4-alpha (HNF-4-alpha) (Transcription factor HNF-4) (Transcription factor 14). | |||||
|
RXRB_HUMAN
|
||||||
| NC score | 0.971308 (rank : 20) | θ value | 2.3998e-69 (rank : 13) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | P28702, P28703 | Gene names | RXRB, NR2B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-beta (Retinoid X receptor beta). | |||||
|
ERR2_MOUSE
|
||||||
| NC score | 0.955353 (rank : 21) | θ value | 1.04822e-48 (rank : 21) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q61539, O09067, O09146 | Gene names | Esrrb, Err-2, Err2, Nr3b2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2). | |||||
|
ERR3_HUMAN
|
||||||
| NC score | 0.955132 (rank : 22) | θ value | 6.7944e-48 (rank : 23) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P62508, O75454, O96021, Q68DA0, Q6P274, Q6PK28, Q6TS38, Q9R1F3, Q9UNJ4 | Gene names | ESRRG, ERR3, ERRG2, KIAA0832, NR3B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3) (ERR gamma-2). | |||||
|
ERR3_MOUSE
|
||||||
| NC score | 0.955132 (rank : 23) | θ value | 6.7944e-48 (rank : 24) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P62509, O75454, O96021, Q8CHC9, Q9R1F3 | Gene names | Esrrg, Err3, Kiaa0832, Nr3b3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Estrogen-related receptor gamma (Estrogen receptor-related protein 3). | |||||
|
ERR2_HUMAN
|
||||||
| NC score | 0.952977 (rank : 24) | θ value | 1.36902e-48 (rank : 22) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O95718, Q9HCB4 | Gene names | ESRRB, ERRB2, NR3B2 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR2 (Estrogen-related receptor, beta) (ERR- beta) (Estrogen receptor-like 2) (ERR beta-2). | |||||
|
ERR1_HUMAN
|
||||||
| NC score | 0.952375 (rank : 25) | θ value | 1.56636e-44 (rank : 25) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | P11474, Q14514 | Gene names | ESRRA, ERR1, ESRL1, NR3B1 | |||
|
Domain Architecture |

|
|||||
| Description | Steroid hormone receptor ERR1 (Estrogen-related receptor, alpha) (ERR- alpha) (Estrogen receptor-like 1). | |||||
|
NR4A2_MOUSE
|
||||||
| NC score | 0.939035 (rank : 26) | θ value | 7.27602e-42 (rank : 27) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q06219, O08690 | Gene names | Nr4a2, Nurr1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (NUR- related factor 1). | |||||
|
NR4A2_HUMAN
|
||||||
| NC score | 0.938994 (rank : 27) | θ value | 5.57106e-42 (rank : 26) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P43354, Q16311 | Gene names | NR4A2, NOT, NURR1, TINUR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A2 (Orphan nuclear receptor NURR1) (Immediate-early response protein NOT) (Transcriptionally-inducible nuclear receptor). | |||||
|
NR4A1_HUMAN
|
||||||
| NC score | 0.938676 (rank : 28) | θ value | 1.79215e-40 (rank : 28) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P22736 | Gene names | NR4A1, GFRP1, HMR, NAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Early response protein NAK1) (TR3 orphan receptor) (ST-59). | |||||
|
NR4A1_MOUSE
|
||||||
| NC score | 0.937378 (rank : 29) | θ value | 3.99248e-40 (rank : 31) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P12813 | Gene names | Nr4a1, Gfrp, Hmr, N10, Nur77 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A1 (Orphan nuclear receptor HMR) (Nuclear hormone receptor NUR/77) (N10 nuclear protein). | |||||
|
NR4A3_MOUSE
|
||||||
| NC score | 0.936514 (rank : 30) | θ value | 2.86122e-38 (rank : 38) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9QZB6, Q9QZB5 | Gene names | Nr4a3, Tec | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Orphan nuclear receptor TEC) (Translocated in extraskeletal chondrosarcoma). | |||||
|
NR4A3_HUMAN
|
||||||
| NC score | 0.935238 (rank : 31) | θ value | 5.96599e-36 (rank : 39) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q92570, Q12935, Q14979, Q16420, Q4VXA9, Q9UEK2, Q9UEK3 | Gene names | NR4A3, CHN, CSMF, MINOR, NOR1, TEC | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Nuclear hormone receptor NOR-1) (Neuron-derived orphan receptor 1) (Mitogen-induced nuclear orphan receptor). | |||||
|
TR4_HUMAN
|
||||||
| NC score | 0.933955 (rank : 32) | θ value | 1.01765e-35 (rank : 40) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P49116, P55092 | Gene names | NR2C2, TAK1, TR4 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor TR4 (Orphan nuclear receptor TAK1). | |||||
|
TR4_MOUSE
|
||||||
| NC score | 0.933597 (rank : 33) | θ value | 1.32909e-35 (rank : 41) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P49117, P55093 | Gene names | Nr2c2, Mtr2r1, Tak1, Tr4 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor TR4 (Orphan nuclear receptor TAK1). | |||||
|
RARA_HUMAN
|
||||||
| NC score | 0.933179 (rank : 34) | θ value | 4.41421e-39 (rank : 35) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P10276, P78456, Q13440, Q13441, Q9NQS0 | Gene names | RARA, NR1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARG2_HUMAN
|
||||||
| NC score | 0.933165 (rank : 35) | θ value | 2.34062e-40 (rank : 30) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P22932, Q15281 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid receptor gamma-2 (RAR-gamma-2). | |||||
|
RARA_MOUSE
|
||||||
| NC score | 0.933084 (rank : 36) | θ value | 7.52953e-39 (rank : 36) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P11416, P22603 | Gene names | Rara, Nr1b1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor alpha (RAR-alpha). | |||||
|
RARG1_HUMAN
|
||||||
| NC score | 0.933020 (rank : 37) | θ value | 2.34062e-40 (rank : 29) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P13631 | Gene names | RARG, NR1B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-1 (RAR-gamma-1). | |||||
|
RARB_HUMAN
|
||||||
| NC score | 0.932631 (rank : 38) | θ value | 7.52953e-39 (rank : 37) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P10826, P12891, Q00989, Q15298, Q9UN48 | Gene names | RARB, HAP, NR1B2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor beta (RAR-beta) (RAR-epsilon) (HBV-activated protein). | |||||
|
RARG2_MOUSE
|
||||||
| NC score | 0.932626 (rank : 39) | θ value | 1.98146e-39 (rank : 34) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P20787 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-B (RAR-gamma-B). | |||||
|
RARB_MOUSE
|
||||||
| NC score | 0.932591 (rank : 40) | θ value | 1.98146e-39 (rank : 32) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P22605, P11417, P22604 | Gene names | Rarb, Nr1b2 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor beta (RAR-beta). | |||||
|
RARG1_MOUSE
|
||||||
| NC score | 0.932336 (rank : 41) | θ value | 1.98146e-39 (rank : 33) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P18911, Q91VK5 | Gene names | Rarg, Nr1b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor gamma-A (RAR-gamma-A). | |||||
|
ESR2_HUMAN
|
||||||
| NC score | 0.932195 (rank : 42) | θ value | 3.74554e-30 (rank : 53) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q92731, O60608, O60685, O60702, O60703, O75583, O75584, Q86Z31, Q9UEV6, Q9UHD3, Q9UQK9 | Gene names | ESR2, ESTRB, NR3A2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
TR2_MOUSE
|
||||||
| NC score | 0.930973 (rank : 43) | θ value | 1.16434e-31 (rank : 46) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q505F1, P97763, Q3UIJ7, Q4U1Z4, Q60927, Q62152, Q8VIJ3 | Gene names | Nr2c1, Tr2, Tr2-11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Orphan nuclear receptor TR2 (Testicular receptor 2) (mTR2). | |||||
|
ESR2_MOUSE
|
||||||
| NC score | 0.930822 (rank : 44) | θ value | 1.6813e-30 (rank : 50) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O08537, O35635, O70519 | Gene names | Esr2, Estrb, Nr3a2 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor beta (ER-beta). | |||||
|
TR2_HUMAN
|
||||||
| NC score | 0.928689 (rank : 45) | θ value | 1.57365e-28 (rank : 57) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P13056 | Gene names | NR2C1, TR2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor TR2 (Testicular receptor 2). | |||||
|
ESR1_MOUSE
|
||||||
| NC score | 0.928561 (rank : 46) | θ value | 2.34606e-32 (rank : 43) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P19785, Q9JJT5, Q9QY51, Q9QY52 | Gene names | Esr1, Esr, Estr, Estra, Nr3a1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
ESR1_HUMAN
|
||||||
| NC score | 0.928482 (rank : 47) | θ value | 8.06329e-33 (rank : 42) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P03372, Q13511, Q14276, Q9NU51, Q9UDZ7, Q9UIS7 | Gene names | ESR1, ESR, NR3A1 | |||
|
Domain Architecture |

|
|||||
| Description | Estrogen receptor (ER) (Estradiol receptor) (ER-alpha). | |||||
|
NR6A1_HUMAN
|
||||||
| NC score | 0.926638 (rank : 48) | θ value | 1.86321e-21 (rank : 87) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q15406, O00551, O00603, Q8NHQ0, Q92898, Q99802 | Gene names | NR6A1, GCNF | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR6A1 (Germ cell nuclear factor) (GCNF) (Retinoid receptor-related testis-specific receptor) (RTR). | |||||
|
NR6A1_MOUSE
|
||||||
| NC score | 0.925559 (rank : 49) | θ value | 3.75424e-22 (rank : 80) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q64249 | Gene names | Nr6a1, Gcnf | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR6A1 (Germ cell nuclear factor) (GCNF) (Retinoid receptor-related testis-specific receptor) (RTR). | |||||
|
NR1H3_HUMAN
|
||||||
| NC score | 0.923489 (rank : 50) | θ value | 5.22648e-32 (rank : 44) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q13133, Q96H87 | Gene names | NR1H3, LXRA | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-alpha (Liver X receptor alpha) (Nuclear orphan receptor LXR-alpha). | |||||
|
NR1H3_MOUSE
|
||||||
| NC score | 0.922248 (rank : 51) | θ value | 1.16434e-31 (rank : 45) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9Z0Y9, Q9QUH7 | Gene names | Nr1h3, Lxra | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-alpha (Liver X receptor alpha) (Nuclear orphan receptor LXR-alpha). | |||||
|
NR1I3_HUMAN
|
||||||
| NC score | 0.922187 (rank : 52) | θ value | 1.12775e-26 (rank : 62) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q14994 | Gene names | NR1I3, CAR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1I3 (Constitutive androstane receptor) (CAR) (Constitutive activator of retinoid response) (Constitutive active response) (Orphan nuclear receptor MB67). | |||||
|
NR5A2_MOUSE
|
||||||
| NC score | 0.921678 (rank : 53) | θ value | 3.50572e-28 (rank : 58) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P45448 | Gene names | Nr5a2, Lrh1 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR5A2 (Liver receptor homolog 1) (LRH-1). | |||||
|
NR1H4_HUMAN
|
||||||
| NC score | 0.921368 (rank : 54) | θ value | 7.80994e-28 (rank : 59) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q96RI1, Q8NFP5, Q8NFP6, Q92943 | Gene names | NR1H4, BAR, FXR, HRR1, RIP14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
NR1H4_MOUSE
|
||||||
| NC score | 0.920658 (rank : 55) | θ value | 5.40856e-29 (rank : 56) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q60641, Q60642, Q60643 | Gene names | Nr1h4, Bar, Fxr, Rip14 | |||
|
Domain Architecture |

|
|||||
| Description | Bile acid receptor (Farnesoid X-activated receptor) (Farnesol receptor HRR-1) (Retinoid X receptor-interacting protein 14) (RXR-interacting protein 14). | |||||
|
NR5A2_HUMAN
|
||||||
| NC score | 0.918438 (rank : 56) | θ value | 4.28545e-26 (rank : 63) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O00482, O95642 | Gene names | NR5A2, B1F, CPF, FTF | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR5A2 (Alpha-1-fetoprotein transcription factor) (Hepatocytic transcription factor) (B1-binding factor) (hB1F) (CYP7A promoter-binding factor) (Liver receptor homolog 1) (LRH-1). | |||||
|
NR1I3_MOUSE
|
||||||
| NC score | 0.917588 (rank : 57) | θ value | 1.05554e-24 (rank : 72) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O35627, O35628 | Gene names | Nr1i3, Car | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1I3 (Constitutive androstane receptor) (CAR) (Orphan nuclear receptor MB67). | |||||
|
PPARA_HUMAN
|
||||||
| NC score | 0.915841 (rank : 58) | θ value | 9.8567e-31 (rank : 48) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q07869, Q16241, Q92486, Q92689, Q9Y3N1 | Gene names | PPARA, NR1C1, PPAR | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor alpha (PPAR-alpha). | |||||
|
STF1_MOUSE
|
||||||
| NC score | 0.915578 (rank : 59) | θ value | 1.42661e-21 (rank : 85) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P33242 | Gene names | Nr5a1, Ftzf1 | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic factor 1 (STF-1) (SF-1) (Adrenal 4-binding protein) (Steroid hormone receptor Ad4BP) (Fushi tarazu factor homolog 1) (Embryonal LTR-binding protein) (ELP) (Embryonal long terminal repeat- binding protein) (Steroid hydroxylase positive regulator). | |||||
|
PPARA_MOUSE
|
||||||
| NC score | 0.914873 (rank : 60) | θ value | 4.89182e-30 (rank : 54) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P23204 | Gene names | Ppara, Nr1c1, Ppar | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor alpha (PPAR-alpha). | |||||
|
PPARD_HUMAN
|
||||||
| NC score | 0.914488 (rank : 61) | θ value | 1.28732e-30 (rank : 49) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q03181, Q7Z5K0, Q9BUD4 | Gene names | PPARD, NR1C2, PPARB | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor delta (PPAR-delta) (PPAR- beta) (Nuclear hormone receptor 1) (NUC1) (NUCI). | |||||
|
NR1H2_MOUSE
|
||||||
| NC score | 0.913820 (rank : 62) | θ value | 1.08979e-29 (rank : 55) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q60644 | Gene names | Nr1h2, Lxrb, Rip15, Unr, Unr2 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Retinoid X receptor-interacting protein No.15). | |||||
|
PPARD_MOUSE
|
||||||
| NC score | 0.913146 (rank : 63) | θ value | 5.77852e-31 (rank : 47) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P35396, P37239 | Gene names | Ppard, Nr1c2, Pparb | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor delta (PPAR-delta) (PPAR- beta) (Nuclear hormone receptor 1) (NUC1). | |||||
|
STF1_HUMAN
|
||||||
| NC score | 0.912876 (rank : 64) | θ value | 1.42661e-21 (rank : 84) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q13285, O15196 | Gene names | NR5A1, AD4BP, FTZF1, SF1 | |||
|
Domain Architecture |

|
|||||
| Description | Steroidogenic factor 1 (STF-1) (SF-1) (Adrenal 4-binding protein) (Steroid hormone receptor Ad4BP) (Fushi tarazu factor homolog 1). | |||||
|
PPARG_HUMAN
|
||||||
| NC score | 0.911906 (rank : 65) | θ value | 2.86786e-30 (rank : 52) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P37231, O00684, O00710, O14515, Q15178, Q15179, Q15180, Q15832, Q86U60, Q96J12 | Gene names | PPARG, NR1C3 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma (PPAR-gamma). | |||||
|
NR1D1_HUMAN
|
||||||
| NC score | 0.911609 (rank : 66) | θ value | 1.42661e-21 (rank : 83) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P20393, Q15304 | Gene names | NR1D1, EAR1, HREV, THRAL | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D1 (V-erbA-related protein EAR-1) (Rev- erbA-alpha). | |||||
|
PPARG_MOUSE
|
||||||
| NC score | 0.911535 (rank : 67) | θ value | 2.19584e-30 (rank : 51) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P37238 | Gene names | Pparg, Nr1c3 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome proliferator-activated receptor gamma (PPAR-gamma). | |||||
|
THA_HUMAN
|
||||||
| NC score | 0.909449 (rank : 68) | θ value | 1.33217e-27 (rank : 60) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P10827, P21205, Q8N6A1, Q96H73 | Gene names | THRA, ERBA1, NR1A1, THRA1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor alpha (C-erbA-alpha) (c-erbA-1) (EAR-7) (EAR7). | |||||
|
THA_MOUSE
|
||||||
| NC score | 0.909443 (rank : 69) | θ value | 1.33217e-27 (rank : 61) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P63058, P10685, P15827, P16416, P37241, Q63107, Q63195, Q63196, Q99146 | Gene names | Thra, C-erba-alpha, Nr1a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor alpha (C-erbA-alpha) (c-erbA-1). | |||||
|
NR1H2_HUMAN
|
||||||
| NC score | 0.905507 (rank : 70) | θ value | 1.86321e-21 (rank : 86) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P55055, Q12970 | Gene names | NR1H2, LXRB, NER, UNR | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterols receptor LXR-beta (Liver X receptor beta) (Nuclear orphan receptor LXR-beta) (Ubiquitously-expressed nuclear receptor) (Nuclear receptor NER). | |||||
|
NR1D2_HUMAN
|
||||||
| NC score | 0.904800 (rank : 71) | θ value | 1.09232e-21 (rank : 82) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q14995, O00402, Q86XD4 | Gene names | NR1D2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (EAR-1R) (Orphan nuclear hormone receptor BD73). | |||||
|
THB2_HUMAN
|
||||||
| NC score | 0.903702 (rank : 72) | θ value | 8.08199e-25 (rank : 71) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P37243 | Gene names | THRB, ERBA2, NR1A2, THR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor beta-2. | |||||
|
THB1_HUMAN
|
||||||
| NC score | 0.903274 (rank : 73) | θ value | 8.08199e-25 (rank : 70) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P10828, Q13986 | Gene names | THRB, ERBA2, NR1A2, THR1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-1. | |||||
|
NR1D2_MOUSE
|
||||||
| NC score | 0.902566 (rank : 74) | θ value | 3.75424e-22 (rank : 79) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q60674, Q60646 | Gene names | Nr1d2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (Orphan nuclear receptor RVR). | |||||
|
RORG_MOUSE
|
||||||
| NC score | 0.902022 (rank : 75) | θ value | 2.12685e-25 (rank : 65) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P51450, Q61027, Q91YT5, Q9QXD9, Q9R177 | Gene names | Rorc, Nr1f3, Rorg, Thor | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-gamma (Nuclear receptor RZR-gamma) (Thymus orphan receptor) (TOR). | |||||
|
THB1_MOUSE
|
||||||
| NC score | 0.901910 (rank : 76) | θ value | 5.23862e-24 (rank : 73) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P37242 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-1. | |||||
|
THB2_MOUSE
|
||||||
| NC score | 0.901774 (rank : 77) | θ value | 5.23862e-24 (rank : 74) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P37244 | Gene names | Thrb, C-erba-beta, Nr1a2 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor beta-2. | |||||
|
RORB_HUMAN
|
||||||
| NC score | 0.899256 (rank : 78) | θ value | 4.73814e-25 (rank : 68) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q92753, Q8WX73 | Gene names | RORB, NR1F2, RZRB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-beta (Nuclear receptor RZR-beta). | |||||
|
RORB_MOUSE
|
||||||
| NC score | 0.899028 (rank : 79) | θ value | 4.73814e-25 (rank : 69) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8R1B8 | Gene names | Rorb, Nr1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-beta (Nuclear receptor RZR-beta). | |||||
|
RORG_HUMAN
|
||||||
| NC score | 0.898792 (rank : 80) | θ value | 1.24688e-25 (rank : 64) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P51449, Q8N5V7, Q8NCY8 | Gene names | RORC, NR1F3, RORG, RZRG | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-gamma (Nuclear receptor RZR-gamma). | |||||
|
RORA_HUMAN
|
||||||
| NC score | 0.897906 (rank : 81) | θ value | 3.62785e-25 (rank : 66) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P35398, P35397, P35399, P45445, Q96H83 | Gene names | RORA, NR1F1, RZRA | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-alpha (Nuclear receptor RZR-alpha). | |||||
|
RORA_MOUSE
|
||||||
| NC score | 0.897877 (rank : 82) | θ value | 3.62785e-25 (rank : 67) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P51448, P70283, P97741, P97773, Q923G1 | Gene names | Rora, Nr1f1, Rzra | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor ROR-alpha (Nuclear receptor RZR-alpha). | |||||
|
VDR_MOUSE
|
||||||
| NC score | 0.897309 (rank : 83) | θ value | 2.13179e-17 (rank : 94) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P48281 | Gene names | Vdr, Nr1i1 | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
VDR_HUMAN
|
||||||
| NC score | 0.895799 (rank : 84) | θ value | 2.13179e-17 (rank : 93) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P11473 | Gene names | VDR, NR1I1 | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin D3 receptor (VDR) (1,25-dihydroxyvitamin D3 receptor). | |||||
|
PXR_MOUSE
|
||||||
| NC score | 0.894664 (rank : 85) | θ value | 3.07829e-16 (rank : 96) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O54915 | Gene names | Nr1i2, Pxr | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor PXR (Pregnane X receptor). | |||||
|
PXR_HUMAN
|
||||||
| NC score | 0.892596 (rank : 86) | θ value | 1.38178e-16 (rank : 95) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | O75469, Q96AC7, Q9UJ22, Q9UJ23, Q9UJ24, Q9UJ25, Q9UJ26, Q9UJ27 | Gene names | NR1I2, PXR | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor PXR (Pregnane X receptor) (Orphan nuclear receptor PAR1) (Steroid and xenobiotic receptor) (SXR). | |||||
|
GCR_HUMAN
|
||||||
| NC score | 0.880433 (rank : 87) | θ value | 3.39556e-23 (rank : 76) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P04150, P04151, Q53EP5, Q6N0A4 | Gene names | NR3C1, GRL | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor (GR). | |||||
|
ANDR_MOUSE
|
||||||
| NC score | 0.877868 (rank : 88) | θ value | 7.56453e-23 (rank : 78) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P19091 | Gene names | Ar, Nr3c4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
ANDR_HUMAN
|
||||||
| NC score | 0.876678 (rank : 89) | θ value | 3.39556e-23 (rank : 75) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | P10275 | Gene names | AR, DHTR, NR3C4 | |||
|
Domain Architecture |

|
|||||
| Description | Androgen receptor (Dihydrotestosterone receptor). | |||||
|
GCR_MOUSE
|
||||||
| NC score | 0.873392 (rank : 90) | θ value | 4.58923e-20 (rank : 91) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P06537, Q61628, Q61629 | Gene names | Nr3c1, Grl, Grl1 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid receptor (GR). | |||||
|
MCR_HUMAN
|
||||||
| NC score | 0.871854 (rank : 91) | θ value | 4.43474e-23 (rank : 77) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | P08235, Q96KQ8, Q96KQ9 | Gene names | NR3C2, MCR, MLR | |||
|
Domain Architecture |

|
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
PRGR_HUMAN
|
||||||
| NC score | 0.869424 (rank : 92) | θ value | 1.86321e-21 (rank : 88) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P06401, Q9UPF7 | Gene names | PGR, NR3C3 | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.868120 (rank : 93) | θ value | 8.36355e-22 (rank : 81) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
PRGR_MOUSE
|
||||||
| NC score | 0.864218 (rank : 94) | θ value | 3.17815e-21 (rank : 89) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q00175 | Gene names | Pgr, Nr3c3, Pr | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone receptor (PR). | |||||
|
SHP_HUMAN
|
||||||
| NC score | 0.646185 (rank : 95) | θ value | 2.69047e-20 (rank : 90) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q15466, Q5QP36 | Gene names | NR0B2, SHP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor 0B2 (Orphan nuclear receptor SHP) (Small heterodimer partner). | |||||
|
SHP_MOUSE
|
||||||
| NC score | 0.639195 (rank : 96) | θ value | 2.5182e-18 (rank : 92) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q62227, Q53Z53 | Gene names | Nr0b2, Shp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor 0B2 (Orphan nuclear receptor SHP) (Small heterodimer partner). | |||||
|
DAX1_MOUSE
|
||||||
| NC score | 0.610337 (rank : 97) | θ value | 6.00763e-12 (rank : 98) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61066, O09068, O09147 | Gene names | Nr0b1, Ahch, Dax1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor 0B1 (Nuclear receptor DAX-1). | |||||
|
DAX1_HUMAN
|
||||||
| NC score | 0.592779 (rank : 98) | θ value | 6.41864e-14 (rank : 97) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P51843, Q96F69 | Gene names | NR0B1, AHC, DAX1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor 0B1 (Nuclear receptor DAX-1) (DSS-AHC critical region on the X chromosome protein 1). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.052274 (rank : 99) | θ value | 0.163984 (rank : 106) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
CO5A2_HUMAN
|
||||||
| NC score | 0.045797 (rank : 100) | θ value | 0.00665767 (rank : 100) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P05997 | Gene names | COL5A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(V) chain precursor. | |||||
|
CO4A2_HUMAN
|
||||||
| NC score | 0.045550 (rank : 101) | θ value | 0.00509761 (rank : 99) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P08572 | Gene names | COL4A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO9A2_HUMAN
|
||||||
| NC score | 0.045328 (rank : 102) | θ value | 0.62314 (rank : 110) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.045117 (rank : 103) | θ value | 0.0961366 (rank : 103) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO1A2_HUMAN
|
||||||
| NC score | 0.044432 (rank : 104) | θ value | 0.21417 (rank : 107) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P08123, P02464, Q9UEB6, Q9UPH0 | Gene names | COL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.044376 (rank : 105) | θ value | 0.0252991 (rank : 101) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO2A1_HUMAN
|
||||||
| NC score | 0.043200 (rank : 106) | θ value | 0.813845 (rank : 111) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P02458, Q12985, Q14009, Q14044, Q14046, Q14056, Q14058, Q16672, Q6LBY1, Q6LBY2, Q6LBY3, Q99227, Q9UE38, Q9UE39, Q9UE40, Q9UE41, Q9UE42, Q9UE43 | Gene names | COL2A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.042718 (rank : 107) | θ value | 1.81305 (rank : 118) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
CO2A1_MOUSE
|
||||||
| NC score | 0.042605 (rank : 108) | θ value | 0.813845 (rank : 112) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P28481 | Gene names | Col2a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(II) chain precursor [Contains: Chondrocalcin]. | |||||
|
CO4A4_HUMAN
|
||||||
| NC score | 0.042249 (rank : 109) | θ value | 0.813845 (rank : 113) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P53420 | Gene names | COL4A4 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-4(IV) chain precursor. | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.041657 (rank : 110) | θ value | 2.36792 (rank : 121) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.040676 (rank : 111) | θ value | 0.163984 (rank : 105) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.040536 (rank : 112) | θ value | 0.21417 (rank : 108) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.039016 (rank : 113) | θ value | 0.365318 (rank : 109) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO4A6_HUMAN
|
||||||
| NC score | 0.038984 (rank : 114) | θ value | 5.27518 (rank : 138) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q14031, Q12823, Q14053, Q5JYH6, Q9NQM5, Q9NTX3, Q9UJ76, Q9UMG6, Q9Y4L4 | Gene names | COL4A6 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-6(IV) chain precursor. | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.038333 (rank : 115) | θ value | 2.36792 (rank : 120) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
CO1A1_HUMAN
|
||||||
| NC score | 0.038097 (rank : 116) | θ value | 4.03905 (rank : 128) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P02452, P78441, Q13896, Q13902, Q13903, Q14037, Q14992, Q15176, Q15201, Q16050, Q7KZ30, Q7KZ34, Q8IVI5, Q9UML6, Q9UMM7 | Gene names | COL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(I) chain precursor. | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.037892 (rank : 117) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CONA1_MOUSE
|
||||||
| NC score | 0.037743 (rank : 118) | θ value | 4.03905 (rank : 133) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8K4G2, Q5SUQ0 | Gene names | Col23a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.037163 (rank : 119) | θ value | 4.03905 (rank : 129) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
COBA1_HUMAN
|
||||||
| NC score | 0.035815 (rank : 120) | θ value | 6.88961 (rank : 144) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P12107, Q14034, Q9UIT4, Q9UIT5, Q9UIT6 | Gene names | COL11A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
C1QC_MOUSE
|
||||||
| NC score | 0.035642 (rank : 121) | θ value | 0.125558 (rank : 104) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q02105 | Gene names | C1qc, C1qg | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1q subcomponent subunit C precursor. | |||||
|
CONA1_HUMAN
|
||||||
| NC score | 0.034671 (rank : 122) | θ value | 4.03905 (rank : 132) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
COAA1_MOUSE
|
||||||
| NC score | 0.034564 (rank : 123) | θ value | 4.03905 (rank : 130) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q05306 | Gene names | Col10a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(X) chain precursor. | |||||
|
COBA1_MOUSE
|
||||||
| NC score | 0.033610 (rank : 124) | θ value | 6.88961 (rank : 145) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 487 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61245, Q64047 | Gene names | Col11a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XI) chain precursor. | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.033535 (rank : 125) | θ value | 3.0926 (rank : 124) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
CO6A1_HUMAN
|
||||||
| NC score | 0.033083 (rank : 126) | θ value | 1.38821 (rank : 116) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
CO6A1_MOUSE
|
||||||
| NC score | 0.032565 (rank : 127) | θ value | 6.88961 (rank : 143) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q04857 | Gene names | Col6a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.032413 (rank : 128) | θ value | 6.88961 (rank : 141) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CO7A1_HUMAN
|
||||||
| NC score | 0.029203 (rank : 129) | θ value | 0.0563607 (rank : 102) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q02388, Q14054, Q16507 | Gene names | COL7A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VII) chain precursor (Long-chain collagen) (LC collagen). | |||||
|
C1QT5_MOUSE
|
||||||
| NC score | 0.029093 (rank : 130) | θ value | 6.88961 (rank : 142) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K479 | Gene names | C1qtnf5, Ctrp5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1q tumor necrosis factor-related protein 5 precursor. | |||||
|
ADIPO_HUMAN
|
||||||
| NC score | 0.023424 (rank : 131) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q15848 | Gene names | ADIPOQ, ACDC, ACRP30, APM1, GBP28 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipose most abundant gene transcript 1) (apM-1) (Gelatin-binding protein). | |||||
|
ADIPO_MOUSE
|
||||||
| NC score | 0.023168 (rank : 132) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q60994, Q62400, Q9DC68 | Gene names | Adipoq, Acdc, Acrp30, Apm1 | |||
|
Domain Architecture |

|
|||||
| Description | Adiponectin precursor (Adipocyte, C1q and collagen domain-containing protein) (30 kDa adipocyte complement-related protein) (Adipocyte complement-related 30 kDa protein) (ACRP30) (Adipocyte-specific protein AdipoQ). | |||||
|
COEA1_HUMAN
|
||||||
| NC score | 0.022107 (rank : 133) | θ value | 4.03905 (rank : 131) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q05707, O00260, O00261, O00262, Q05708, Q5XJ18, Q96C67 | Gene names | COL14A1, UND | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XIV) chain precursor (Undulin). | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.019901 (rank : 134) | θ value | 4.03905 (rank : 137) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.015545 (rank : 135) | θ value | 1.06291 (rank : 114) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
FCN1_MOUSE
|
||||||
| NC score | 0.013301 (rank : 136) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O70165 | Gene names | Fcn1, Fcna | |||
|
Domain Architecture |

|
|||||
| Description | Ficolin-1 precursor (Collagen/fibrinogen domain-containing protein 1) (Ficolin-A) (Ficolin A) (M-Ficolin). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.010330 (rank : 137) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RMD5B_MOUSE
|
||||||
| NC score | 0.009709 (rank : 138) | θ value | 6.88961 (rank : 148) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YQ7 | Gene names | Rmnd5b | |||
|
Domain Architecture |

|
|||||
| Description | RMD5 homolog B. | |||||
|
RMD5B_HUMAN
|
||||||
| NC score | 0.008723 (rank : 139) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96G75, Q6UVY7, Q9H6F6 | Gene names | RMND5B | |||
|
Domain Architecture |

|
|||||
| Description | RMD5 homolog B. | |||||
|
ERB1_HUMAN
|
||||||
| NC score | 0.008220 (rank : 140) | θ value | 6.88961 (rank : 146) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P60509 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-R(b)_3p24.3 provirus ancestral Env polyprotein precursor (Envelope polyprotein) (HERV-R(b) Env protein) [Includes: Surface protein domain (SU); Transmembrane protein domain (TM)]. | |||||
|
PODO_HUMAN
|
||||||
| NC score | 0.007063 (rank : 141) | θ value | 4.03905 (rank : 136) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NP85, Q8N6Q5 | Gene names | NPHS2 | |||
|
Domain Architecture |

|
|||||
| Description | Podocin. | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.006677 (rank : 142) | θ value | 2.36792 (rank : 123) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
CXCC1_HUMAN
|
||||||
| NC score | 0.006553 (rank : 143) | θ value | 3.0926 (rank : 125) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
RTEL1_HUMAN
|
||||||
| NC score | 0.005490 (rank : 144) | θ value | 3.0926 (rank : 127) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZ71, Q5JTV4, Q9BW37, Q9H402, Q9H4X6, Q9NX25, Q9NZ73, Q9UPR4, Q9Y4R6 | Gene names | RTEL1, C20orf41, KIAA1088, NHL | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of telomere elongation helicase 1 (EC 3.6.1.-) (Helicase- like protein NHL). | |||||
|
SI1L1_HUMAN
|
||||||
| NC score | 0.004868 (rank : 145) | θ value | 5.27518 (rank : 139) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43166, O95321, Q9UDU4, Q9UNU4 | Gene names | SIPA1L1, E6TP1, KIAA0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1 (High-risk human papilloma viruses E6 oncoproteins targeted protein 1) (E6- targeted protein 1). | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.004576 (rank : 146) | θ value | 3.0926 (rank : 126) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
CXCC1_MOUSE
|
||||||
| NC score | 0.004285 (rank : 147) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.004153 (rank : 148) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.002046 (rank : 149) | θ value | 4.03905 (rank : 135) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
KCNH6_HUMAN
|
||||||
| NC score | 0.001753 (rank : 150) | θ value | 4.03905 (rank : 134) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H252, Q9BRD7 | Gene names | KCNH6, ERG2, HERG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 6 (Voltage-gated potassium channel subunit Kv11.2) (Ether-a-go-go-related gene potassium channel 2) (Ether-a-go-go-related protein 2) (Eag-related protein 2). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.000996 (rank : 151) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.000600 (rank : 152) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | -0.000563 (rank : 153) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
BOC_HUMAN
|
||||||
| NC score | -0.000786 (rank : 154) | θ value | 2.36792 (rank : 119) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BWV1, Q6UXJ5, Q8N2P7, Q8NF26 | Gene names | BOC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brother of CDO precursor (Protein BOC). | |||||
|
INVS_MOUSE
|
||||||
| NC score | -0.002787 (rank : 155) | θ value | 2.36792 (rank : 122) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
ZN341_HUMAN
|
||||||
| NC score | -0.005559 (rank : 156) | θ value | 5.27518 (rank : 140) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9BYN7, Q5JXM8, Q96ST5 | Gene names | ZNF341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 341. | |||||
|
ZN335_HUMAN
|
||||||
| NC score | -0.006372 (rank : 157) | θ value | 1.38821 (rank : 117) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ULK1_HUMAN
|
||||||
| NC score | -0.007345 (rank : 158) | θ value | 1.06291 (rank : 115) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
PKN3_MOUSE
|
||||||
| NC score | -0.008416 (rank : 159) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1009 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K045 | Gene names | Pkn3, Pknbeta | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase N3 (EC 2.7.11.13) (Protein kinase PKN- beta) (Protein-kinase C-related kinase 3). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | -0.008597 (rank : 160) | θ value | 6.88961 (rank : 147) | |||
| Query Neighborhood Hits | 160 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||