Please be patient as the page loads
|
SIRT1_MOUSE
|
||||||
| SwissProt Accessions | Q923E4, Q9QXG8 | Gene names | Sirt1, Sir2l1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (SIR2alpha) (mSIR2a) (Sir2) (SIR2-like protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SIRT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991419 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96EB6, Q2XNF6, Q5JVQ0, Q9GZR9, Q9Y6F0 | Gene names | SIRT1, SIR2L1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (hSIRT1) (hSIR2) (SIR2-like protein 1). | |||||
|
SIRT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q923E4, Q9QXG8 | Gene names | Sirt1, Sir2l1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (SIR2alpha) (mSIR2a) (Sir2) (SIR2-like protein 1). | |||||
|
SIRT2_MOUSE
|
||||||
| θ value | 1.51013e-55 (rank : 3) | NC score | 0.906312 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VDQ8, Q9CXS5, Q9EQ18, Q9ERJ9 | Gene names | Sirt2, Sir2l2 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-2 (EC 3.5.1.-) (SIR2-like protein 2) (mSIR2L2). | |||||
|
SIRT2_HUMAN
|
||||||
| θ value | 4.11246e-53 (rank : 4) | NC score | 0.903581 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXJ6, O95889, Q924Y7, Q9P0G8, Q9UNT0, Q9Y6E9 | Gene names | SIRT2, SIR2L, SIR2L2 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-2 (EC 3.5.1.-) (SIR2-like) (SIR2- like protein 2). | |||||
|
SIRT3_HUMAN
|
||||||
| θ value | 2.75849e-49 (rank : 5) | NC score | 0.903688 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NTG7, Q9Y6E8 | Gene names | SIRT3, SIR2L3 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-3, mitochondrial precursor (EC 3.5.1.-) (SIR2-like protein 3) (hSIRT3). | |||||
|
SIRT3_MOUSE
|
||||||
| θ value | 2.26182e-43 (rank : 6) | NC score | 0.902608 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R104, Q9EPA8 | Gene names | Sirt3, Sir2l3 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-3 (EC 3.5.1.-) (SIR2-like protein 3) (mSIR2L3). | |||||
|
SIRT5_HUMAN
|
||||||
| θ value | 2.97466e-19 (rank : 7) | NC score | 0.725411 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NXA8, Q9Y6E6 | Gene names | SIRT5, SIR2L5 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
SIRT4_MOUSE
|
||||||
| θ value | 8.10077e-17 (rank : 8) | NC score | 0.699954 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R216 | Gene names | Sirt4, Sir2l4 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-4 (EC 3.5.1.-) (SIR2-like protein 4). | |||||
|
SIRT5_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 9) | NC score | 0.716122 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2C6 | Gene names | Sirt5, Sir2l5 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
SIRT4_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 10) | NC score | 0.697516 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6E7, O43346 | Gene names | SIRT4, SIR2L4 | |||
|
Domain Architecture |
|
|||||
| Description | NAD-dependent deacetylase sirtuin-4 (EC 3.5.1.-) (SIR2-like protein 4). | |||||
|
SIRT6_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 11) | NC score | 0.619091 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N6T7, O75291, Q6IAF5, Q6PK99, Q8NCD2, Q9BSI5, Q9BWP3, Q9NRC7, Q9UQD1 | Gene names | SIRT6, SIR2L6 | |||
|
Domain Architecture |
|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
SIRT6_MOUSE
|
||||||
| θ value | 2.52405e-10 (rank : 12) | NC score | 0.603922 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59941 | Gene names | Sirt6, Sir2l6 | |||
|
Domain Architecture |
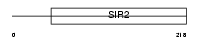
|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
SIRT7_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 13) | NC score | 0.516524 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BKJ9, Q6X7B7, Q8QZZ5 | Gene names | Sirt7, Sir2l7 | |||
|
Domain Architecture |
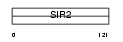
|
|||||
| Description | NAD-dependent deacetylase sirtuin-7 (EC 3.5.1.-) (SIR2-like protein 7). | |||||
|
SIRT7_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 14) | NC score | 0.505603 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRC8, Q3MIK4, Q9NSZ6, Q9NUS6 | Gene names | SIRT7, SIR2L7 | |||
|
Domain Architecture |
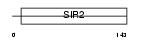
|
|||||
| Description | NAD-dependent deacetylase sirtuin-7 (EC 3.5.1.-) (SIR2-like protein 7). | |||||
|
L2GL1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.030867 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15334, O00188, Q58F11, Q86UK6 | Gene names | LLGL1, HUGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 1 (LLGL) (Human homolog to the D-lgl gene) (Hugl-1) (DLG4). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.009946 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CD2L2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.002451 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
PHF8_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.020833 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TJ7, Q8BLX8, Q8BLY0, Q8BZ61, Q8CG26 | Gene names | Phf8, Kiaa1111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
CPSF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.039026 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
CPSF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.039080 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.021208 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.006522 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.019642 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
NRK_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.002284 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
DDX51_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.008298 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.012305 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RIMB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.014423 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.015979 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.010256 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.017095 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
DOC10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.009194 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
ITCH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.005187 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
LRRK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.000837 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
RNF25_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.016137 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZR0, Q3TAL8, Q9DCW7 | Gene names | Rnf25 | |||
|
Domain Architecture |
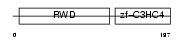
|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-) (RING finger protein AO7). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.009175 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.012559 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CD36_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.010449 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08857 | Gene names | Cd36 | |||
|
Domain Architecture |
|
|||||
| Description | Platelet glycoprotein 4 (Platelet glycoprotein IV) (GPIV) (GPIIIB) (CD36 antigen) (PAS IV) (PAS-4 protein). | |||||
|
ELF4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.006295 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.015328 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.010256 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
REPS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.011517 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RTN4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.010608 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
SCN3A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.005972 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.005752 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.005988 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.019130 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MAGA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.007319 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |
|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
NEK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | -0.000314 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |
|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||
|
PHF8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.015571 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
SCN2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.007159 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SIRT1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q923E4, Q9QXG8 | Gene names | Sirt1, Sir2l1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (SIR2alpha) (mSIR2a) (Sir2) (SIR2-like protein 1). | |||||
|
SIRT1_HUMAN
|
||||||
| NC score | 0.991419 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96EB6, Q2XNF6, Q5JVQ0, Q9GZR9, Q9Y6F0 | Gene names | SIRT1, SIR2L1 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-1 (EC 3.5.1.-) (hSIRT1) (hSIR2) (SIR2-like protein 1). | |||||
|
SIRT2_MOUSE
|
||||||
| NC score | 0.906312 (rank : 3) | θ value | 1.51013e-55 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8VDQ8, Q9CXS5, Q9EQ18, Q9ERJ9 | Gene names | Sirt2, Sir2l2 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-2 (EC 3.5.1.-) (SIR2-like protein 2) (mSIR2L2). | |||||
|
SIRT3_HUMAN
|
||||||
| NC score | 0.903688 (rank : 4) | θ value | 2.75849e-49 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NTG7, Q9Y6E8 | Gene names | SIRT3, SIR2L3 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-3, mitochondrial precursor (EC 3.5.1.-) (SIR2-like protein 3) (hSIRT3). | |||||
|
SIRT2_HUMAN
|
||||||
| NC score | 0.903581 (rank : 5) | θ value | 4.11246e-53 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IXJ6, O95889, Q924Y7, Q9P0G8, Q9UNT0, Q9Y6E9 | Gene names | SIRT2, SIR2L, SIR2L2 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-2 (EC 3.5.1.-) (SIR2-like) (SIR2- like protein 2). | |||||
|
SIRT3_MOUSE
|
||||||
| NC score | 0.902608 (rank : 6) | θ value | 2.26182e-43 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R104, Q9EPA8 | Gene names | Sirt3, Sir2l3 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-3 (EC 3.5.1.-) (SIR2-like protein 3) (mSIR2L3). | |||||
|
SIRT5_HUMAN
|
||||||
| NC score | 0.725411 (rank : 7) | θ value | 2.97466e-19 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NXA8, Q9Y6E6 | Gene names | SIRT5, SIR2L5 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
SIRT5_MOUSE
|
||||||
| NC score | 0.716122 (rank : 8) | θ value | 4.02038e-16 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K2C6 | Gene names | Sirt5, Sir2l5 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-5 (EC 3.5.1.-) (SIR2-like protein 5). | |||||
|
SIRT4_MOUSE
|
||||||
| NC score | 0.699954 (rank : 9) | θ value | 8.10077e-17 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R216 | Gene names | Sirt4, Sir2l4 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-4 (EC 3.5.1.-) (SIR2-like protein 4). | |||||
|
SIRT4_HUMAN
|
||||||
| NC score | 0.697516 (rank : 10) | θ value | 4.44505e-15 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6E7, O43346 | Gene names | SIRT4, SIR2L4 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-4 (EC 3.5.1.-) (SIR2-like protein 4). | |||||
|
SIRT6_HUMAN
|
||||||
| NC score | 0.619091 (rank : 11) | θ value | 1.33837e-11 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N6T7, O75291, Q6IAF5, Q6PK99, Q8NCD2, Q9BSI5, Q9BWP3, Q9NRC7, Q9UQD1 | Gene names | SIRT6, SIR2L6 | |||
|
Domain Architecture |

|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
SIRT6_MOUSE
|
||||||
| NC score | 0.603922 (rank : 12) | θ value | 2.52405e-10 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P59941 | Gene names | Sirt6, Sir2l6 | |||
|
Domain Architecture |

|
|||||
| Description | Mono-ADP-ribosyltransferase sirtuin-6 (EC 2.4.2.31) (SIR2-like protein 6). | |||||
|
SIRT7_MOUSE
|
||||||
| NC score | 0.516524 (rank : 13) | θ value | 1.43324e-05 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8BKJ9, Q6X7B7, Q8QZZ5 | Gene names | Sirt7, Sir2l7 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-7 (EC 3.5.1.-) (SIR2-like protein 7). | |||||
|
SIRT7_HUMAN
|
||||||
| NC score | 0.505603 (rank : 14) | θ value | 0.000121331 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRC8, Q3MIK4, Q9NSZ6, Q9NUS6 | Gene names | SIRT7, SIR2L7 | |||
|
Domain Architecture |

|
|||||
| Description | NAD-dependent deacetylase sirtuin-7 (EC 3.5.1.-) (SIR2-like protein 7). | |||||
|
CPSF1_MOUSE
|
||||||
| NC score | 0.039080 (rank : 15) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EPU4 | Gene names | Cpsf1, Cpsf160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
CPSF1_HUMAN
|
||||||
| NC score | 0.039026 (rank : 16) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q10570, Q96AF0 | Gene names | CPSF1, CPSF160 | |||
|
Domain Architecture |

|
|||||
| Description | Cleavage and polyadenylation specificity factor 160 kDa subunit (CPSF 160 kDa subunit). | |||||
|
L2GL1_HUMAN
|
||||||
| NC score | 0.030867 (rank : 17) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15334, O00188, Q58F11, Q86UK6 | Gene names | LLGL1, HUGL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lethal(2) giant larvae protein homolog 1 (LLGL) (Human homolog to the D-lgl gene) (Hugl-1) (DLG4). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.021208 (rank : 18) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PHF8_MOUSE
|
||||||
| NC score | 0.020833 (rank : 19) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80TJ7, Q8BLX8, Q8BLY0, Q8BZ61, Q8CG26 | Gene names | Phf8, Kiaa1111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.019642 (rank : 20) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.019130 (rank : 21) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.017095 (rank : 22) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
RNF25_MOUSE
|
||||||
| NC score | 0.016137 (rank : 23) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QZR0, Q3TAL8, Q9DCW7 | Gene names | Rnf25 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 25 (EC 6.3.2.-) (RING finger protein AO7). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.015979 (rank : 24) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.015571 (rank : 25) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.015328 (rank : 26) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
RIMB1_HUMAN
|
||||||
| NC score | 0.014423 (rank : 27) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1479 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95153, O75111, Q8N5W3 | Gene names | BZRAP1, KIAA0612, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.012559 (rank : 28) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.012305 (rank : 29) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
REPS1_MOUSE
|
||||||
| NC score | 0.011517 (rank : 30) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O54916, Q8C9J9, Q99LR8 | Gene names | Reps1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
RTN4_HUMAN
|
||||||
| NC score | 0.010608 (rank : 31) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQC3, O94962, Q9BXG5, Q9H212, Q9H3I3, Q9UQ42, Q9Y293, Q9Y2Y7, Q9Y5U6 | Gene names | RTN4, KIAA0886, NOGO | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein) (Foocen) (Neuroendocrine-specific protein) (NSP) (Neuroendocrine-specific protein C homolog) (RTN-x) (Reticulon-5). | |||||
|
CD36_MOUSE
|
||||||
| NC score | 0.010449 (rank : 32) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08857 | Gene names | Cd36 | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein 4 (Platelet glycoprotein IV) (GPIV) (GPIIIB) (CD36 antigen) (PAS IV) (PAS-4 protein). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.010256 (rank : 33) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.010256 (rank : 34) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.009946 (rank : 35) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
DOC10_HUMAN
|
||||||
| NC score | 0.009194 (rank : 36) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96BY6, O75178, Q9NW06, Q9NXI8 | Gene names | DOCK10, KIAA0694 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 10 (Protein zizimin 3). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.009175 (rank : 37) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
DDX51_HUMAN
|
||||||
| NC score | 0.008298 (rank : 38) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N8A6, Q5CZ71, Q8IXK5, Q96ED1 | Gene names | DDX51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX51 (EC 3.6.1.-) (DEAD box protein 51). | |||||
|
MAGA5_HUMAN
|
||||||
| NC score | 0.007319 (rank : 39) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43359 | Gene names | MAGEA5, MAGE5 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen 5 (MAGE-5 antigen). | |||||
|
SCN2A_HUMAN
|
||||||
| NC score | 0.007159 (rank : 40) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99250, Q14472, Q9BZC9, Q9BZD0 | Gene names | SCN2A, NAC2, SCN2A2 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 2 subunit alpha (Sodium channel protein type II subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.2) (Sodium channel protein, brain II subunit alpha) (HBSC II). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.006522 (rank : 41) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
ELF4_MOUSE
|
||||||
| NC score | 0.006295 (rank : 42) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2U4 | Gene names | Elf4, Mef | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-4 (E74-like factor 4) (Myeloid Elf-1-like factor). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.005988 (rank : 43) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SCN3A_HUMAN
|
||||||
| NC score | 0.005972 (rank : 44) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NY46, Q16142, Q9BZB3, Q9C006, Q9NYK2, Q9UPD1, Q9Y6P4 | Gene names | SCN3A, NAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium channel protein type 3 subunit alpha (Sodium channel protein type III subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.3) (Sodium channel protein, brain III subunit alpha) (Voltage- gated sodium channel subtype III). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.005752 (rank : 45) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
ITCH_HUMAN
|
||||||
| NC score | 0.005187 (rank : 46) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96J02, O43584, Q5TEL0, Q96F66, Q9BY75, Q9H451, Q9H4U5 | Gene names | ITCH | |||
|
Domain Architecture |

|
|||||
| Description | Itchy homolog E3 ubiquitin protein ligase (EC 6.3.2.-) (Itch) (Atrophin-1-interacting protein 4) (AIP4) (NFE2-associated polypeptide 1) (NAPP1). | |||||
|
CD2L2_HUMAN
|
||||||
| NC score | 0.002451 (rank : 47) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1439 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UQ88, O95227, O95228, O96012, Q12821, Q12853, Q12854, Q5QPR0, Q5QPR1, Q5QPR2, Q9UBC4, Q9UBI3, Q9UEI1, Q9UEI2, Q9UP53, Q9UP54, Q9UP55, Q9UP56, Q9UQ86, Q9UQ87, Q9UQ89 | Gene names | CDC2L2, PITSLREB | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L2 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 2) (CDK11). | |||||
|
NRK_HUMAN
|
||||||
| NC score | 0.002284 (rank : 48) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z2Y5, Q32ND6, Q5H9K2, Q6ZMP2 | Gene names | NRK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nik-related protein kinase (EC 2.7.11.1). | |||||
|
LRRK1_MOUSE
|
||||||
| NC score | 0.000837 (rank : 49) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1267 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHC2, Q3U476, Q66JQ4, Q6GQR9, Q6NZF5, Q6ZPI4, Q8BKP3, Q8BU93, Q8BUY0, Q8BVV2, Q8R085 | Gene names | Lrrk1, Kiaa1790 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 1 (EC 2.7.11.1). | |||||
|
NEK1_HUMAN
|
||||||
| NC score | -0.000314 (rank : 50) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1129 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96PY6, Q9Y594 | Gene names | NEK1, KIAA1901 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1) (NY-REN-55 antigen). | |||||