Please be patient as the page loads
|
PERF_MOUSE
|
||||||
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PERF_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.976970 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14222, Q86WX7 | Gene names | PRF1, PFP | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (PFP) (Cytolysin). | |||||
|
PERF_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
|
CO6_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 3) | NC score | 0.404149 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
CO8A_MOUSE
|
||||||
| θ value | 8.38298e-14 (rank : 4) | NC score | 0.443990 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K182, Q8CHJ9, Q8CHK1 | Gene names | C8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
CO8A_HUMAN
|
||||||
| θ value | 8.67504e-11 (rank : 5) | NC score | 0.424507 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07357, Q13668, Q9H130 | Gene names | C8A | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
CO7_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 6) | NC score | 0.374584 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
CO8B_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 7) | NC score | 0.386640 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH35, Q4VAH1, Q8CHJ5, Q8CHJ6, Q8VC14 | Gene names | C8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
CO9_MOUSE
|
||||||
| θ value | 1.53129e-07 (rank : 8) | NC score | 0.422343 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06683, Q91XA7 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor. | |||||
|
CO9_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 9) | NC score | 0.401896 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02748 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor [Contains: Complement component C9a; Complement component C9b]. | |||||
|
CO8B_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 10) | NC score | 0.370050 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07358 | Gene names | C8B | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
RASL1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 11) | NC score | 0.162951 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
RASL1_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 12) | NC score | 0.157495 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
FA62A_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 13) | NC score | 0.167398 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
FA62A_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 14) | NC score | 0.161814 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
UN13B_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 15) | NC score | 0.152678 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
OTOF_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.157301 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
OTOF_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 17) | NC score | 0.157700 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
MYO10_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 18) | NC score | 0.020405 (rank : 55) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
UN13A_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 19) | NC score | 0.146185 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.139070 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
DYSF_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 21) | NC score | 0.132434 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
MYOF_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.128080 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
RASL2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.109857 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.027566 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.027598 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
K0528_HUMAN
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.098442 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.098584 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.017999 (rank : 58) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
FAM5A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.083556 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60477, Q6IPV6, Q6P1A0, Q8WU22 | Gene names | FAM5A, DBC1, DBCCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5A precursor (Deleted in bladder cancer 1 protein). | |||||
|
FAM5A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.083462 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q920P3, Q80ZL2, Q812F0, Q8C1C9, Q920P4, Q9QXL0 | Gene names | Fam5a, Brinp, Brinp1, Dbccr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5A precursor (Deleted in bladder cancer 1 protein homolog) (BMP/retinoic acid-inducible neural-specific protein 1). | |||||
|
FAM5B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.084712 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6DFY8, Q80T96 | Gene names | Fam5b, Kiaa1747 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5B precursor. | |||||
|
DOC2A_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.056673 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
FAM5B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.082604 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0B6, O95560, Q6ZWC1, Q7LCZ9, Q8N360 | Gene names | FAM5B, BRINP2, DBCCR1L2, KIAA1747 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5B precursor (BMP/retinoic acid-inducible neural-specific protein 2) (DBCCR1-like protein 2). | |||||
|
KPCA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.019007 (rank : 56) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
SYLM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.026042 (rank : 53) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
CPNE4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.033781 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |
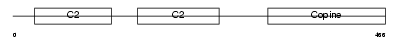
|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.033689 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
FAM5C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.073757 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76B58, O95726 | Gene names | FAM5C, DBCCR1L, DBCCR1L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor (DBCCR1-like protein 1). | |||||
|
FAM5C_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.074216 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q499E0, Q8K0R9 | Gene names | Fam5c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor. | |||||
|
CIZ1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.022004 (rank : 54) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
TR11B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.014410 (rank : 59) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00300, O60236, Q53FX6, Q9UHP4 | Gene names | TNFRSF11B, OCIF, OPG | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
CPNE1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.036640 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |
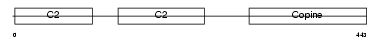
|
|||||
| Description | Copine-1 (Copine I). | |||||
|
DOC2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.054052 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
MT3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.041504 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
KPCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.018102 (rank : 57) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
BAIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.051479 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
CR001_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.055599 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
Domain Architecture |
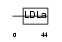
|
|||||
| Description | Protein C18orf1. | |||||
|
CR001_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.055294 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
DOC2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.052891 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DOC2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.050530 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
DYSF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.106451 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
RASA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.062464 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.063796 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.062179 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RASA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.061435 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
RFIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.054785 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
RP3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.052619 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
RP3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.053924 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
UN13D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.057684 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
PERF_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P10820 | Gene names | Prf1, Pfp | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (Cytolysin). | |||||
|
PERF_HUMAN
|
||||||
| NC score | 0.976970 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P14222, Q86WX7 | Gene names | PRF1, PFP | |||
|
Domain Architecture |

|
|||||
| Description | Perforin-1 precursor (P1) (Lymphocyte pore-forming protein) (PFP) (Cytolysin). | |||||
|
CO8A_MOUSE
|
||||||
| NC score | 0.443990 (rank : 3) | θ value | 8.38298e-14 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K182, Q8CHJ9, Q8CHK1 | Gene names | C8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
CO8A_HUMAN
|
||||||
| NC score | 0.424507 (rank : 4) | θ value | 8.67504e-11 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07357, Q13668, Q9H130 | Gene names | C8A | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 alpha chain precursor (Complement component 8 subunit alpha). | |||||
|
CO9_MOUSE
|
||||||
| NC score | 0.422343 (rank : 5) | θ value | 1.53129e-07 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P06683, Q91XA7 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor. | |||||
|
CO6_HUMAN
|
||||||
| NC score | 0.404149 (rank : 6) | θ value | 4.02038e-16 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P13671 | Gene names | C6 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C6 precursor. | |||||
|
CO9_HUMAN
|
||||||
| NC score | 0.401896 (rank : 7) | θ value | 2.61198e-07 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P02748 | Gene names | C9 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C9 precursor [Contains: Complement component C9a; Complement component C9b]. | |||||
|
CO8B_MOUSE
|
||||||
| NC score | 0.386640 (rank : 8) | θ value | 2.36244e-08 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BH35, Q4VAH1, Q8CHJ5, Q8CHJ6, Q8VC14 | Gene names | C8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
CO7_HUMAN
|
||||||
| NC score | 0.374584 (rank : 9) | θ value | 1.38499e-08 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10643, Q6P3T5, Q92489 | Gene names | C7 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C7 precursor. | |||||
|
CO8B_HUMAN
|
||||||
| NC score | 0.370050 (rank : 10) | θ value | 2.44474e-05 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P07358 | Gene names | C8B | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C8 beta chain precursor (Complement component 8 subunit beta). | |||||
|
FA62A_HUMAN
|
||||||
| NC score | 0.167398 (rank : 11) | θ value | 0.000786445 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BSJ8, O94848, Q6PJN4, Q9H6J1, Q9H6W2, Q9Y416 | Gene names | FAM62A, KIAA0747, MBC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
RASL1_HUMAN
|
||||||
| NC score | 0.162951 (rank : 12) | θ value | 0.00020696 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O95294, Q52M03, Q96CC7 | Gene names | RASAL1, RASAL | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
FA62A_MOUSE
|
||||||
| NC score | 0.161814 (rank : 13) | θ value | 0.00390308 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3U7R1, Q8C8R1, Q91X62, Q9CVH0, Q9Z1X5, Q9Z1X6 | Gene names | Fam62a, Mbc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM62A (Membrane-bound C2 domain-containing protein). | |||||
|
OTOF_MOUSE
|
||||||
| NC score | 0.157700 (rank : 14) | θ value | 0.0193708 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESF1, Q9ESF2 | Gene names | Otof, Fer1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
RASL1_MOUSE
|
||||||
| NC score | 0.157495 (rank : 15) | θ value | 0.000602161 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z268 | Gene names | Rasal1, Rasal | |||
|
Domain Architecture |

|
|||||
| Description | RasGAP-activating-like protein 1. | |||||
|
OTOF_HUMAN
|
||||||
| NC score | 0.157301 (rank : 16) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HC10, Q9HC08, Q9HC09, Q9Y650 | Gene names | OTOF, FER1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Otoferlin (Fer-1-like protein 2). | |||||
|
UN13B_MOUSE
|
||||||
| NC score | 0.152678 (rank : 17) | θ value | 0.00869519 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z1N9 | Gene names | Unc13b, Unc13a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
UN13A_HUMAN
|
||||||
| NC score | 0.146185 (rank : 18) | θ value | 0.0252991 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPW8 | Gene names | UNC13A, KIAA1032 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog A (Munc13-1). | |||||
|
UN13B_HUMAN
|
||||||
| NC score | 0.139070 (rank : 19) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14795 | Gene names | UNC13B, UNC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog B (Munc13-2) (munc13). | |||||
|
DYSF_HUMAN
|
||||||
| NC score | 0.132434 (rank : 20) | θ value | 0.0563607 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75923, O75696, Q9UEN7 | Gene names | DYSF, FER1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
MYOF_HUMAN
|
||||||
| NC score | 0.128080 (rank : 21) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZM1, Q9HBU3, Q9NZM0, Q9ULL3, Q9Y4U4 | Gene names | FER1L3, KIAA1207, MYOF | |||
|
Domain Architecture |

|
|||||
| Description | Myoferlin (Fer-1-like protein 3). | |||||
|
RASL2_HUMAN
|
||||||
| NC score | 0.109857 (rank : 22) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43374, O60286, Q86UW3, Q96QU0 | Gene names | RASA4, CAPRI, GAPL, KIAA0538 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2) (Calcium-promoted Ras inactivator). | |||||
|
DYSF_MOUSE
|
||||||
| NC score | 0.106451 (rank : 23) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESD7, Q9QXC0 | Gene names | Dysf, Fer1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Dysferlin (Dystrophy-associated fer-1-like protein) (Fer-1-like protein 1). | |||||
|
K0528_MOUSE
|
||||||
| NC score | 0.098584 (rank : 24) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7TPS5, Q3TQY6, Q6A052, Q80VA1, Q8C0U3, Q8CID5, Q9CS85 | Gene names | Kiaa0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
K0528_HUMAN
|
||||||
| NC score | 0.098442 (rank : 25) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86YS7, O60280, Q7Z619, Q86SU3 | Gene names | KIAA0528 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0528. | |||||
|
FAM5B_MOUSE
|
||||||
| NC score | 0.084712 (rank : 26) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6DFY8, Q80T96 | Gene names | Fam5b, Kiaa1747 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5B precursor. | |||||
|
FAM5A_HUMAN
|
||||||
| NC score | 0.083556 (rank : 27) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60477, Q6IPV6, Q6P1A0, Q8WU22 | Gene names | FAM5A, DBC1, DBCCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5A precursor (Deleted in bladder cancer 1 protein). | |||||
|
FAM5A_MOUSE
|
||||||
| NC score | 0.083462 (rank : 28) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q920P3, Q80ZL2, Q812F0, Q8C1C9, Q920P4, Q9QXL0 | Gene names | Fam5a, Brinp, Brinp1, Dbccr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5A precursor (Deleted in bladder cancer 1 protein homolog) (BMP/retinoic acid-inducible neural-specific protein 1). | |||||
|
FAM5B_HUMAN
|
||||||
| NC score | 0.082604 (rank : 29) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0B6, O95560, Q6ZWC1, Q7LCZ9, Q8N360 | Gene names | FAM5B, BRINP2, DBCCR1L2, KIAA1747 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5B precursor (BMP/retinoic acid-inducible neural-specific protein 2) (DBCCR1-like protein 2). | |||||
|
FAM5C_MOUSE
|
||||||
| NC score | 0.074216 (rank : 30) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q499E0, Q8K0R9 | Gene names | Fam5c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor. | |||||
|
FAM5C_HUMAN
|
||||||
| NC score | 0.073757 (rank : 31) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q76B58, O95726 | Gene names | FAM5C, DBCCR1L, DBCCR1L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5C precursor (DBCCR1-like protein 1). | |||||
|
RASA2_MOUSE
|
||||||
| NC score | 0.063796 (rank : 32) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P58069 | Gene names | Rasa2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.062464 (rank : 33) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
RASA3_HUMAN
|
||||||
| NC score | 0.062179 (rank : 34) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14644 | Gene names | RASA3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein). | |||||
|
RASA3_MOUSE
|
||||||
| NC score | 0.061435 (rank : 35) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q60790 | Gene names | Rasa3 | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 3 (GAP1(IP4BP)) (Ins P4-binding protein) (GapIII). | |||||
|
UN13D_HUMAN
|
||||||
| NC score | 0.057684 (rank : 36) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q70J99, Q9H7K5 | Gene names | UNC13D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Unc-13 homolog D (Munc13-4). | |||||
|
DOC2A_HUMAN
|
||||||
| NC score | 0.056673 (rank : 37) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14183, Q6P4G4, Q7Z5G0, Q8IVX0 | Gene names | DOC2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha) (Doc2). | |||||
|
CR001_HUMAN
|
||||||
| NC score | 0.055599 (rank : 38) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15165, O15166, O15167, O15168, Q6NXP3 | Gene names | C18orf1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein C18orf1. | |||||
|
CR001_MOUSE
|
||||||
| NC score | 0.055294 (rank : 39) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BWJ4 | Gene names | D18Ertd653e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C18orf1 homolog. | |||||
|
RFIP2_HUMAN
|
||||||
| NC score | 0.054785 (rank : 40) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7L804, Q9Y2F0 | Gene names | RAB11FIP2, KIAA0941 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 2 (Rab11-FIP2) (NRip11). | |||||
|
DOC2A_MOUSE
|
||||||
| NC score | 0.054052 (rank : 41) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TNF0 | Gene names | Doc2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein alpha (Doc2-alpha). | |||||
|
RP3A_MOUSE
|
||||||
| NC score | 0.053924 (rank : 42) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P47708 | Gene names | Rph3a | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
DOC2B_HUMAN
|
||||||
| NC score | 0.052891 (rank : 43) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14184 | Gene names | DOC2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
RP3A_HUMAN
|
||||||
| NC score | 0.052619 (rank : 44) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y2J0, Q96AE0 | Gene names | RPH3A, KIAA0985 | |||
|
Domain Architecture |

|
|||||
| Description | Rabphilin-3A (Exophilin-1). | |||||
|
BAIP3_HUMAN
|
||||||
| NC score | 0.051479 (rank : 45) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O94812, O94839, Q2M226, Q658J2, Q76N05, Q96RZ3, Q9UJK1 | Gene names | BAIAP3, KIAA0734 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BAI1-associated protein 3 (BAP3). | |||||
|
DOC2B_MOUSE
|
||||||
| NC score | 0.050530 (rank : 46) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70169, Q6NXK3 | Gene names | Doc2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double C2-like domain-containing protein beta (Doc2-beta). | |||||
|
MT3_HUMAN
|
||||||
| NC score | 0.041504 (rank : 47) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P25713, Q2V574 | Gene names | MT3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF) (GIFB). | |||||
|
CPNE1_MOUSE
|
||||||
| NC score | 0.036640 (rank : 48) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8C166, Q925K4, Q925K5 | Gene names | Cpne1 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-1 (Copine I). | |||||
|
CPNE4_HUMAN
|
||||||
| NC score | 0.033781 (rank : 49) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96A23, Q8TEX1 | Gene names | CPNE4 | |||
|
Domain Architecture |

|
|||||
| Description | Copine-4 (Copine IV) (Copine-8). | |||||
|
CPNE4_MOUSE
|
||||||
| NC score | 0.033689 (rank : 50) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BLR2 | Gene names | Cpne4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Copine-4 (Copine IV). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.027598 (rank : 51) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.027566 (rank : 52) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
SYLM_HUMAN
|
||||||
| NC score | 0.026042 (rank : 53) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15031 | Gene names | LARS2, KIAA0028 | |||
|
Domain Architecture |

|
|||||
| Description | Probable leucyl-tRNA synthetase, mitochondrial precursor (EC 6.1.1.4) (Leucine--tRNA ligase) (LeuRS). | |||||
|
CIZ1_HUMAN
|
||||||
| NC score | 0.022004 (rank : 54) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULV3, Q9NYM8, Q9UHK4, Q9Y3F9, Q9Y3G0 | Gene names | CIZ1, LSFR1, NP94 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cip1-interacting zinc finger protein (Nuclear protein NP94). | |||||
|
MYO10_HUMAN
|
||||||
| NC score | 0.020405 (rank : 55) | θ value | 0.0252991 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 920 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HD67, Q9NYM7, Q9P110, Q9P111, Q9UHF6 | Gene names | MYO10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin X). | |||||
|
KPCA_MOUSE
|
||||||
| NC score | 0.019007 (rank : 56) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1067 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20444 | Gene names | Prkca, Pkca | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
KPCA_HUMAN
|
||||||
| NC score | 0.018102 (rank : 57) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1070 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P17252, Q15137, Q96RE4 | Gene names | PRKCA, PKCA, PRKACA | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C alpha type (EC 2.7.11.13) (PKC-alpha) (PKC-A). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.017999 (rank : 58) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
TR11B_HUMAN
|
||||||
| NC score | 0.014410 (rank : 59) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00300, O60236, Q53FX6, Q9UHP4 | Gene names | TNFRSF11B, OCIF, OPG | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||