Please be patient as the page loads
|
ING2_HUMAN
|
||||||
| SwissProt Accessions | Q9H160, O95698 | Gene names | ING2, ING1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein) (ING1Lp) (p32). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ING2_MOUSE
|
||||||
| θ value | 2.00348e-116 (rank : 1) | NC score | 0.991681 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9ESK4, Q80VI5, Q8BGU8 | Gene names | Ing2, Ing1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein). | |||||
|
ING2_HUMAN
|
||||||
| θ value | 1.34386e-112 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9H160, O95698 | Gene names | ING2, ING1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein) (ING1Lp) (p32). | |||||
|
ING1_MOUSE
|
||||||
| θ value | 1.22964e-73 (rank : 3) | NC score | 0.971634 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QXV3, Q9QUP8, Q9QXV4, Q9QZX3 | Gene names | Ing1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING1_HUMAN
|
||||||
| θ value | 1.36585e-56 (rank : 4) | NC score | 0.955413 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UK53, O00532, O43658, Q9H007, Q9HD98, Q9HD99, Q9NS83, Q9P0U6, Q9UBC6, Q9UIJ1, Q9UIJ2, Q9UIJ3, Q9UIJ4, Q9UK52 | Gene names | ING1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING5_MOUSE
|
||||||
| θ value | 6.15952e-41 (rank : 5) | NC score | 0.910159 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
ING5_HUMAN
|
||||||
| θ value | 1.6774e-38 (rank : 6) | NC score | 0.910656 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WYH8, Q9BS30 | Gene names | ING5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5 (p28ING5). | |||||
|
ING4_MOUSE
|
||||||
| θ value | 2.67802e-36 (rank : 7) | NC score | 0.907098 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING4_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 8) | NC score | 0.907847 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING3_HUMAN
|
||||||
| θ value | 1.37858e-24 (rank : 9) | NC score | 0.793956 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
ING3_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 10) | NC score | 0.789984 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
INGX_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 11) | NC score | 0.905329 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P0U5 | Gene names | INGX, ING2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein, X-linked (Inhibitor of growth protein 2) (ING1-like tumor suppressor protein). | |||||
|
CXCC1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 12) | NC score | 0.362050 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |
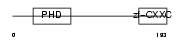
|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 13) | NC score | 0.358415 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 14) | NC score | 0.216087 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 15) | NC score | 0.172529 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 16) | NC score | 0.309161 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 17) | NC score | 0.305562 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 18) | NC score | 0.242830 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 19) | NC score | 0.156919 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 20) | NC score | 0.203314 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 21) | NC score | 0.161986 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 22) | NC score | 0.140444 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 23) | NC score | 0.140398 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
CTGE5_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 24) | NC score | 0.055782 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 25) | NC score | 0.050261 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.056988 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.139439 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
STK10_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 28) | NC score | 0.019301 (rank : 122) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
SH24A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 29) | NC score | 0.063929 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.113663 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 0.21417 (rank : 31) | NC score | 0.125113 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
CCD11_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.054371 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
STK10_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.016915 (rank : 124) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.112870 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.116363 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.109520 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.108368 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.365318 (rank : 38) | NC score | 0.112462 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 39) | NC score | 0.081482 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.053485 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.051771 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 42) | NC score | 0.040371 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.052137 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 44) | NC score | 0.052945 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 45) | NC score | 0.133742 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 46) | NC score | 0.053256 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
NEK9_HUMAN
|
||||||
| θ value | 0.62314 (rank : 47) | NC score | 0.004613 (rank : 130) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.62314 (rank : 48) | NC score | 0.053311 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
AF17_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.118460 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 0.813845 (rank : 50) | NC score | 0.036635 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
PHF8_HUMAN
|
||||||
| θ value | 0.813845 (rank : 51) | NC score | 0.122125 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
PHF8_MOUSE
|
||||||
| θ value | 0.813845 (rank : 52) | NC score | 0.122698 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80TJ7, Q8BLX8, Q8BLY0, Q8BZ61, Q8CG26 | Gene names | Phf8, Kiaa1111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
AF10_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.114756 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.114770 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.044545 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CI093_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.048305 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.039427 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.158420 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.039044 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
HDAC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.020913 (rank : 120) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
SH24A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.052702 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.050175 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.032056 (rank : 108) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.068819 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.025681 (rank : 116) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.032450 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.075960 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.046942 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
MSH2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.024394 (rank : 117) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.159560 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.159031 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.140430 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.043402 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
FOG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.007845 (rank : 128) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
K1718_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.119166 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
K1718_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.118422 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
KIF5A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.029087 (rank : 111) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.028788 (rank : 112) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
MSH2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.021920 (rank : 119) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
RPGR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.040648 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.010496 (rank : 127) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.011480 (rank : 126) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
ANLN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.022329 (rank : 118) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.043865 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.033570 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.071290 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
P55G_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.030507 (rank : 110) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
SAFB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.026687 (rank : 115) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.040400 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
P55G_HUMAN
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.027604 (rank : 114) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
PHF12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.115616 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PLCB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.014427 (rank : 125) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.020171 (rank : 121) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.019068 (rank : 123) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
CDR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.037462 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.038796 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
HS105_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.007674 (rank : 129) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.031137 (rank : 109) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.028121 (rank : 113) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.111298 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.038543 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.117199 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.092090 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.110174 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.073006 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.070056 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.066474 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.063861 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.075312 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.099244 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.068744 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.068850 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JADE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050417 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050448 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JHD1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051802 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2K7, Q49A21, Q4G0M3, Q69YY8, Q9BVH5, Q9H7H5, Q9UK66 | Gene names | FBXL11, FBL7, JHDM1A, KIAA1004 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11) (F-box protein FBL7) (F-box protein Lilina). | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.074062 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.055861 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.052222 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.050445 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051814 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.085636 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.081892 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.061893 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.083141 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.066772 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.081084 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.080311 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.066962 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.067266 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.065899 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ING2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.34386e-112 (rank : 2) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9H160, O95698 | Gene names | ING2, ING1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein) (ING1Lp) (p32). | |||||
|
ING2_MOUSE
|
||||||
| NC score | 0.991681 (rank : 2) | θ value | 2.00348e-116 (rank : 1) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9ESK4, Q80VI5, Q8BGU8 | Gene names | Ing2, Ing1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein). | |||||
|
ING1_MOUSE
|
||||||
| NC score | 0.971634 (rank : 3) | θ value | 1.22964e-73 (rank : 3) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QXV3, Q9QUP8, Q9QXV4, Q9QZX3 | Gene names | Ing1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING1_HUMAN
|
||||||
| NC score | 0.955413 (rank : 4) | θ value | 1.36585e-56 (rank : 4) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UK53, O00532, O43658, Q9H007, Q9HD98, Q9HD99, Q9NS83, Q9P0U6, Q9UBC6, Q9UIJ1, Q9UIJ2, Q9UIJ3, Q9UIJ4, Q9UK52 | Gene names | ING1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING5_HUMAN
|
||||||
| NC score | 0.910656 (rank : 5) | θ value | 1.6774e-38 (rank : 6) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WYH8, Q9BS30 | Gene names | ING5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5 (p28ING5). | |||||
|
ING5_MOUSE
|
||||||
| NC score | 0.910159 (rank : 6) | θ value | 6.15952e-41 (rank : 5) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
ING4_HUMAN
|
||||||
| NC score | 0.907847 (rank : 7) | θ value | 1.46948e-34 (rank : 8) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING4_MOUSE
|
||||||
| NC score | 0.907098 (rank : 8) | θ value | 2.67802e-36 (rank : 7) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
INGX_HUMAN
|
||||||
| NC score | 0.905329 (rank : 9) | θ value | 4.59992e-12 (rank : 11) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9P0U5 | Gene names | INGX, ING2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein, X-linked (Inhibitor of growth protein 2) (ING1-like tumor suppressor protein). | |||||
|
ING3_HUMAN
|
||||||
| NC score | 0.793956 (rank : 10) | θ value | 1.37858e-24 (rank : 9) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
ING3_MOUSE
|
||||||
| NC score | 0.789984 (rank : 11) | θ value | 4.01107e-24 (rank : 10) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
CXCC1_HUMAN
|
||||||
| NC score | 0.362050 (rank : 12) | θ value | 8.40245e-06 (rank : 12) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_MOUSE
|
||||||
| NC score | 0.358415 (rank : 13) | θ value | 1.87187e-05 (rank : 13) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.309161 (rank : 14) | θ value | 0.00134147 (rank : 16) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.305562 (rank : 15) | θ value | 0.00134147 (rank : 17) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.242830 (rank : 16) | θ value | 0.00665767 (rank : 18) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.216087 (rank : 17) | θ value | 0.00035302 (rank : 14) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.203314 (rank : 18) | θ value | 0.00869519 (rank : 20) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.172529 (rank : 19) | θ value | 0.00035302 (rank : 15) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.161986 (rank : 20) | θ value | 0.0113563 (rank : 21) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.159560 (rank : 21) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.159031 (rank : 22) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.158420 (rank : 23) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.156919 (rank : 24) | θ value | 0.00665767 (rank : 19) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.140444 (rank : 25) | θ value | 0.0431538 (rank : 22) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.140430 (rank : 26) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.140398 (rank : 27) | θ value | 0.0431538 (rank : 23) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.139439 (rank : 28) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.133742 (rank : 29) | θ value | 0.62314 (rank : 45) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.125113 (rank : 30) | θ value | 0.21417 (rank : 31) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
PHF8_MOUSE
|
||||||
| NC score | 0.122698 (rank : 31) | θ value | 0.813845 (rank : 52) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q80TJ7, Q8BLX8, Q8BLY0, Q8BZ61, Q8CG26 | Gene names | Phf8, Kiaa1111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
PHF8_HUMAN
|
||||||
| NC score | 0.122125 (rank : 32) | θ value | 0.813845 (rank : 51) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPP1, Q5H9U5, Q5VUJ4, Q7Z6D4, Q9HAH2 | Gene names | PHF8, KIAA1111, ZNF422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 8. | |||||
|
K1718_HUMAN
|
||||||
| NC score | 0.119166 (rank : 33) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZMT4, Q6MZL8, Q9C0E5 | Gene names | KIAA1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.118460 (rank : 34) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
K1718_MOUSE
|
||||||
| NC score | 0.118422 (rank : 35) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q3UWM4, Q3UWN8, Q6ZPJ5, Q8C969, Q8C9E0, Q91VX8 | Gene names | Kiaa1718 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1718. | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.117199 (rank : 36) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.116363 (rank : 37) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.115616 (rank : 38) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.114770 (rank : 39) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.114756 (rank : 40) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.113663 (rank : 41) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.112870 (rank : 42) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.112462 (rank : 43) | θ value | 0.365318 (rank : 38) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.111298 (rank : 44) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.110174 (rank : 45) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.109520 (rank : 46) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.108368 (rank : 47) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.099244 (rank : 48) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.092090 (rank : 49) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.085636 (rank : 50) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.083141 (rank : 51) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.081892 (rank : 52) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.081482 (rank : 53) | θ value | 0.365318 (rank : 39) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.081084 (rank : 54) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.080311 (rank : 55) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.075960 (rank : 56) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.075312 (rank : 57) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.074062 (rank : 58) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.073006 (rank : 59) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.071290 (rank : 60) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.070056 (rank : 61) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.068850 (rank : 62) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.068819 (rank : 63) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.068744 (rank : 64) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.067266 (rank : 65) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.066962 (rank : 66) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.066772 (rank : 67) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.066474 (rank : 68) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.065899 (rank : 69) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
SH24A_HUMAN
|
||||||
| NC score | 0.063929 (rank : 70) | θ value | 0.125558 (rank : 29) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H788, Q5XKC1, Q6NXE9, Q86YM2, Q96C88, Q9H7F7 | Gene names | SH2D4A, SH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A (Protein SH(2)A). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.063861 (rank : 71) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.061893 (rank : 72) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.056988 (rank : 73) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.055861 (rank : 74) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
CTGE5_MOUSE
|
||||||
| NC score | 0.055782 (rank : 75) | θ value | 0.0736092 (rank : 24) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 928 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8R311, Q8CIE3 | Gene names | Ctage5, Mea6, Mgea6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 homolog (cTAGE-5 protein) (Meningioma-expressed antigen 6). | |||||
|
CCD11_HUMAN
|
||||||
| NC score | 0.054371 (rank : 76) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96M91 | Gene names | CCDC11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.053485 (rank : 77) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.053311 (rank : 78) | θ value | 0.62314 (rank : 48) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.053256 (rank : 79) | θ value | 0.62314 (rank : 46) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.052945 (rank : 80) | θ value | 0.62314 (rank : 44) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
SH24A_MOUSE
|
||||||
| NC score | 0.052702 (rank : 81) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.052222 (rank : 82) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.052137 (rank : 83) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.051814 (rank : 84) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
JHD1A_HUMAN
|
||||||
| NC score | 0.051802 (rank : 85) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2K7, Q49A21, Q4G0M3, Q69YY8, Q9BVH5, Q9H7H5, Q9UK66 | Gene names | FBXL11, FBL7, JHDM1A, KIAA1004 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11) (F-box protein FBL7) (F-box protein Lilina). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.051771 (rank : 86) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.050448 (rank : 87) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.050445 (rank : 88) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.050417 (rank : 89) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.050261 (rank : 90) | θ value | 0.0736092 (rank : 25) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.050175 (rank : 91) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.048305 (rank : 92) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.046942 (rank : 93) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.044545 (rank : 94) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.043865 (rank : 95) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.043402 (rank : 96) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
RPGR1_HUMAN
|
||||||
| NC score | 0.040648 (rank : 97) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q96KN7, Q7Z2W6, Q8IXV5, Q96QA8, Q9HB94, Q9HB95, Q9HBK6, Q9NR40 | Gene names | RPGRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.040400 (rank : 98) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.040371 (rank : 99) | θ value | 0.47712 (rank : 42) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.039427 (rank : 100) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.039044 (rank : 101) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.038796 (rank : 102) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.038543 (rank : 103) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CDR2_MOUSE
|
||||||
| NC score | 0.037462 (rank : 104) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P97817 | Gene names | Cdr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebellar degeneration-related protein 2. | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.036635 (rank : 105) | θ value | 0.813845 (rank : 50) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.033570 (rank : 106) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.032450 (rank : 107) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.032056 (rank : 108) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.031137 (rank : 109) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
P55G_MOUSE
|
||||||
| NC score | 0.030507 (rank : 110) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q64143 | Gene names | Pik3r3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
KIF5A_HUMAN
|
||||||
| NC score | 0.029087 (rank : 111) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12840, Q4LE26 | Gene names | KIF5A, NKHC1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.028788 (rank : 112) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.028121 (rank : 113) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
P55G_HUMAN
|
||||||
| NC score | 0.027604 (rank : 114) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q92569, O60482 | Gene names | PIK3R3 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 3-kinase regulatory subunit gamma (PI3-kinase p85-subunit gamma) (PtdIns-3-kinase p85-gamma) (p55PIK). | |||||
|
SAFB2_MOUSE
|
||||||
| NC score | 0.026687 (rank : 115) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80YR5, Q8K153 | Gene names | Safb2 | |||
|
Domain Architecture |

|
|||||
| Description | Scaffold attachment factor B2. | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.025681 (rank : 116) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
MSH2_MOUSE
|
||||||
| NC score | 0.024394 (rank : 117) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.022329 (rank : 118) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MSH2_HUMAN
|
||||||
| NC score | 0.021920 (rank : 119) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
HDAC4_HUMAN
|
||||||
| NC score | 0.020913 (rank : 120) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P56524, Q9UND6 | Gene names | HDAC4, KIAA0288 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 4 (HD4). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.020171 (rank : 121) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
STK10_MOUSE
|
||||||
| NC score | 0.019301 (rank : 122) | θ value | 0.0961366 (rank : 28) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 2132 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O55098 | Gene names | Stk10, Lok | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.019068 (rank : 123) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
STK10_HUMAN
|
||||||
| NC score | 0.016915 (rank : 124) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O94804, Q9UIW4 | Gene names | STK10, LOK | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase 10 (EC 2.7.11.1) (Lymphocyte-oriented kinase). | |||||
|
PLCB3_HUMAN
|
||||||
| NC score | 0.014427 (rank : 125) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.011480 (rank : 126) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.010496 (rank : 127) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
FOG2_HUMAN
|
||||||
| NC score | 0.007845 (rank : 128) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WW38, Q9NPL7, Q9NPS4, Q9UNI5 | Gene names | ZFPM2, FOG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM2 (Zinc finger protein multitype 2) (Friend of GATA protein 2) (FOG-2) (hFOG-2). | |||||
|
HS105_HUMAN
|
||||||
| NC score | 0.007674 (rank : 129) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92598, O95739, Q5TBM6, Q5TBM8, Q9UPC4 | Gene names | HSPH1, HSP105, HSP110, KIAA0201 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein 105 kDa (Heat shock 110 kDa protein) (Antigen NY- CO-25). | |||||
|
NEK9_HUMAN
|
||||||
| NC score | 0.004613 (rank : 130) | θ value | 0.62314 (rank : 47) | |||
| Query Neighborhood Hits | 101 | Target Neighborhood Hits | 900 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8TD19, Q52LK6, Q8NCN0, Q8TCY4, Q9UPI4, Q9Y6S4, Q9Y6S5, Q9Y6S6 | Gene names | NEK9, KIAA1995, NEK8, NERCC | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek9 (EC 2.7.11.1) (NimA-related protein kinase 9) (Never in mitosis A-related kinase 9) (Nercc1 kinase) (NIMA-related kinase 8) (Nek8). | |||||