Please be patient as the page loads
|
CRFR1_HUMAN
|
||||||
| SwissProt Accessions | P34998, Q13008, Q9UK64 | Gene names | CRHR1, CRFR, CRFR1, CRHR | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 1 precursor (CRF-R) (CRF1) (Corticotropin-releasing hormone receptor 1) (CRH-R 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CRFR1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P34998, Q13008, Q9UK64 | Gene names | CRHR1, CRFR, CRFR1, CRHR | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 1 precursor (CRF-R) (CRF1) (Corticotropin-releasing hormone receptor 1) (CRH-R 1). | |||||
|
CRFR1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998116 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35347 | Gene names | Crhr1, Crhr | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 1 precursor (CRF-R) (CRF1) (Corticotropin-releasing hormone receptor 1) (CRH-R 1). | |||||
|
CRFR2_HUMAN
|
||||||
| θ value | 1.56915e-153 (rank : 3) | NC score | 0.994166 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13324, O43461, Q99431 | Gene names | CRHR2, CRF2R, CRH2R | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 2 precursor (CRF-R 2) (CRF2) (Corticotropin-releasing hormone receptor 2) (CRH-R 2). | |||||
|
CRFR2_MOUSE
|
||||||
| θ value | 6.59259e-152 (rank : 4) | NC score | 0.993639 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60748, Q60783, Q60808 | Gene names | Crhr2, Crf2r | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 2 precursor (CRF-R 2) (CRF2) (Corticotropin-releasing hormone receptor 2) (CRH-R 2) (CRF-RB) (CRH- R2). | |||||
|
PTHR1_HUMAN
|
||||||
| θ value | 2.75849e-49 (rank : 5) | NC score | 0.940506 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q03431 | Gene names | PTHR1, PTHR | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor (PTH/PTHr receptor) (PTH/PTHrP type I receptor). | |||||
|
CALCR_HUMAN
|
||||||
| θ value | 7.51208e-47 (rank : 6) | NC score | 0.928724 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P30988, O14585, Q13941, Q6DJU8, Q6T712 | Gene names | CALCR | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin receptor precursor (CT-R). | |||||
|
CALCR_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 7) | NC score | 0.932638 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q60755 | Gene names | Calcr | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin receptor precursor (CT-R). | |||||
|
PTHR1_MOUSE
|
||||||
| θ value | 7.51208e-47 (rank : 8) | NC score | 0.935681 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P41593, Q62119 | Gene names | Pthr1, Pthr | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor (PTH/PTHr receptor) (PTH/PTHrP type I receptor). | |||||
|
VIPR1_MOUSE
|
||||||
| θ value | 3.48949e-44 (rank : 9) | NC score | 0.934966 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P97751, Q9JI40, Q9R1T8 | Gene names | Vipr1 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 1 precursor (VIP-R-1) (VPAC1) (Pituitary adenylate cyclase-activating polypeptide type II receptor) (PACAP type II receptor) (PACAP-R-2). | |||||
|
GLP1R_MOUSE
|
||||||
| θ value | 7.27602e-42 (rank : 10) | NC score | 0.920625 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O35659 | Gene names | Glp1r | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 1 receptor precursor (GLP-1 receptor) (GLP-1-R) (GLP-1R). | |||||
|
PTHR2_MOUSE
|
||||||
| θ value | 9.50281e-42 (rank : 11) | NC score | 0.915309 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q91V95 | Gene names | Pthr2 | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone receptor precursor (PTH2 receptor). | |||||
|
PTHR2_HUMAN
|
||||||
| θ value | 2.11701e-41 (rank : 12) | NC score | 0.918010 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P49190, Q8N429 | Gene names | PTHR2 | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone receptor precursor (PTH2 receptor). | |||||
|
GLP1R_HUMAN
|
||||||
| θ value | 1.05066e-40 (rank : 13) | NC score | 0.915731 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P43220, Q99669 | Gene names | GLP1R | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 1 receptor precursor (GLP-1 receptor) (GLP-1-R) (GLP-1R). | |||||
|
GHRHR_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 14) | NC score | 0.921047 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q02643, Q99863 | Gene names | GHRHR | |||
|
Domain Architecture |

|
|||||
| Description | Growth hormone-releasing hormone receptor precursor (GHRH receptor) (GRF receptor) (GRFR). | |||||
|
VIPR2_MOUSE
|
||||||
| θ value | 8.89434e-40 (rank : 15) | NC score | 0.923629 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P41588, P97750 | Gene names | Vipr2 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 2 precursor (VIP-R-2) (Pituitary adenylate cyclase-activating polypeptide type III receptor) (PACAP type III receptor) (PACAP-R-3). | |||||
|
GLR_HUMAN
|
||||||
| θ value | 3.37985e-39 (rank : 16) | NC score | 0.917361 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P47871 | Gene names | GCGR | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon receptor precursor (GL-R). | |||||
|
VIPR2_HUMAN
|
||||||
| θ value | 5.76516e-39 (rank : 17) | NC score | 0.922420 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P41587, Q13053, Q15870 | Gene names | VIPR2, VIP2R | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 2 precursor (VIP-R-2) (Pituitary adenylate cyclase-activating polypeptide type III receptor) (PACAP type III receptor) (PACAP-R-3) (Helodermin-preferring VIP receptor). | |||||
|
SCTR_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 18) | NC score | 0.918317 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P47872, Q12961, Q13213 | Gene names | SCTR | |||
|
Domain Architecture |

|
|||||
| Description | Secretin receptor precursor (SCT-R). | |||||
|
VIPR1_HUMAN
|
||||||
| θ value | 4.88048e-38 (rank : 19) | NC score | 0.924782 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P32241, Q15871 | Gene names | VIPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 1 precursor (VIP-R-1) (Pituitary adenylate cyclase-activating polypeptide type II receptor) (PACAP type II receptor) (PACAP-R-2). | |||||
|
GLR_MOUSE
|
||||||
| θ value | 2.05049e-36 (rank : 20) | NC score | 0.912999 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61606, Q63960 | Gene names | Gcgr | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon receptor precursor (GL-R). | |||||
|
PACR_MOUSE
|
||||||
| θ value | 1.73584e-35 (rank : 21) | NC score | 0.921725 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70205 | Gene names | Adcyap1r1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary adenylate cyclase-activating polypeptide type I receptor precursor (PACAP type I receptor) (PACAP-R-1). | |||||
|
PACR_HUMAN
|
||||||
| θ value | 1.12514e-34 (rank : 22) | NC score | 0.921387 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P41586 | Gene names | ADCYAP1R1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary adenylate cyclase-activating polypeptide type I receptor precursor (PACAP type I receptor) (PACAP-R-1). | |||||
|
SCTR_MOUSE
|
||||||
| θ value | 4.72714e-33 (rank : 23) | NC score | 0.912664 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5FWI2, Q80T66 | Gene names | Sctr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretin receptor precursor (SCT-R). | |||||
|
CALRL_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 24) | NC score | 0.912372 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q16602 | Gene names | CALCRL, CGRPR | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin gene-related peptide type 1 receptor precursor (CGRP type 1 receptor) (Calcitonin receptor-like receptor). | |||||
|
GLP2R_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 25) | NC score | 0.906727 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95838 | Gene names | GLP2R | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 2 receptor precursor (GLP-2 receptor) (GLP-2-R) (GLP-2R). | |||||
|
CALRL_MOUSE
|
||||||
| θ value | 2.59387e-31 (rank : 26) | NC score | 0.910509 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9R1W5, Q9QXH8, Q9WUP2 | Gene names | Calcrl, Crlr | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin gene-related peptide type 1 receptor precursor (CGRP type 1 receptor) (Calcitonin receptor-like receptor). | |||||
|
GHRHR_MOUSE
|
||||||
| θ value | 4.42448e-31 (rank : 27) | NC score | 0.909072 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P32082 | Gene names | Ghrhr, Grfr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth hormone-releasing hormone receptor precursor (GHRH receptor) (GRF receptor) (GRFR). | |||||
|
GIPR_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 28) | NC score | 0.906646 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48546, Q14401, Q16400, Q9UPI1 | Gene names | GIPR | |||
|
Domain Architecture |

|
|||||
| Description | Gastric inhibitory polypeptide receptor precursor (GIP-R) (Glucose- dependent insulinotropic polypeptide receptor). | |||||
|
GP133_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 29) | NC score | 0.744577 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
EMR4_MOUSE
|
||||||
| θ value | 2.35696e-16 (rank : 30) | NC score | 0.658068 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91ZE5, Q3U1I7, Q8BYX0, Q8VIM3 | Gene names | Emr4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 4 precursor (EGF-like module-containing mucin-like receptor EMR4) (F4/80-like-receptor) (Seven-span membrane protein FIRE). | |||||
|
EMR3_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 31) | NC score | 0.632767 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BY15 | Gene names | EMR3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 3 precursor (EGF-like module-containing mucin-like receptor EMR3). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 1.2105e-12 (rank : 32) | NC score | 0.476604 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 33) | NC score | 0.475824 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
EMR1_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 34) | NC score | 0.451473 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14246, Q8NGA7 | Gene names | EMR1, TM7LN3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor). | |||||
|
GPR64_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 35) | NC score | 0.605862 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
LPHN3_HUMAN
|
||||||
| θ value | 1.02475e-11 (rank : 36) | NC score | 0.574324 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9HAR2, O94867, Q9NWK5 | Gene names | LPHN3, KIAA0768, LEC3 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-3 precursor (Calcium-independent alpha-latrotoxin receptor 3) (Lectomedin-3). | |||||
|
GPR64_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 37) | NC score | 0.611011 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
EMR2_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 38) | NC score | 0.444824 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHX3, Q8NG96, Q9Y4B1 | Gene names | EMR2 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 2 precursor (EGF-like module EMR2) (CD312 antigen). | |||||
|
BAI3_HUMAN
|
||||||
| θ value | 1.133e-10 (rank : 39) | NC score | 0.452948 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O60242, O60297 | Gene names | BAI3, KIAA0550 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
BAI3_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 40) | NC score | 0.451898 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q80ZF8 | Gene names | Bai3 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
CD97_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 41) | NC score | 0.510079 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z0M6, Q923A1, Q9CVI5, Q9JLQ8 | Gene names | Cd97 | |||
|
Domain Architecture |

|
|||||
| Description | CD97 antigen precursor. | |||||
|
EMR1_MOUSE
|
||||||
| θ value | 1.133e-10 (rank : 42) | NC score | 0.466380 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
CD97_HUMAN
|
||||||
| θ value | 2.52405e-10 (rank : 43) | NC score | 0.467976 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P48960, O00718, O76101, Q8NG72, Q8TBQ7 | Gene names | CD97 | |||
|
Domain Architecture |

|
|||||
| Description | CD97 antigen precursor (Leukocyte antigen CD97). | |||||
|
GP128_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 44) | NC score | 0.564211 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96K78, Q86SQ2 | Gene names | GPR128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
GP116_HUMAN
|
||||||
| θ value | 2.79066e-09 (rank : 45) | NC score | 0.467232 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IZF2, O94858, Q6RGN2, Q86SP0, Q9Y3Z2 | Gene names | GPR116, KIAA0758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 116 precursor. | |||||
|
GP115_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 46) | NC score | 0.503169 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D2L6 | Gene names | Gpr115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 115 precursor. | |||||
|
GP110_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 47) | NC score | 0.494409 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q5T601, Q5KU15, Q5T5Z9, Q86SM1, Q8IXE3, Q8IZF8, Q96DQ1 | Gene names | GPR110, PGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 110 precursor (G-protein coupled receptor PGR19) (G-protein coupled receptor KPG_012). | |||||
|
BAI2_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 48) | NC score | 0.453845 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI2_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 49) | NC score | 0.452567 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
GP110_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 50) | NC score | 0.492013 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VEC3, Q8BMV8 | Gene names | Gpr110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 110 precursor. | |||||
|
GP144_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 51) | NC score | 0.645069 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
ELTD1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 52) | NC score | 0.560318 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9HBW9 | Gene names | ELTD1, ETL | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin and seven transmembrane domain-containing protein 1 precursor (EGF-TM7-latrophilin-related protein) (ETL protein). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 53) | NC score | 0.583323 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
LPHN2_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 54) | NC score | 0.549258 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O95490, O94882, Q5VX76, Q9UKY5, Q9UKY6 | Gene names | LPHN2, KIAA0786, LEC1, LPHH1 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-2 precursor (Calcium-independent alpha-latrotoxin receptor 2) (Latrophilin homolog 1) (Lectomedin-1). | |||||
|
ELTD1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 55) | NC score | 0.496811 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q923X1, Q91W44 | Gene names | Eltd1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin seven transmembrane domain-containing protein 1 precursor. | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 56) | NC score | 0.284330 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 57) | NC score | 0.276569 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
GP113_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 58) | NC score | 0.575713 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8IZF5, Q6UXT7, Q6UXX3, Q86SL7, Q8IXD8, Q8TDT3 | Gene names | GPR113, PGR23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 113 precursor (G-protein coupled receptor PGR23). | |||||
|
GP126_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 59) | NC score | 0.483566 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 9.92553e-07 (rank : 60) | NC score | 0.261686 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
GP115_HUMAN
|
||||||
| θ value | 2.88788e-06 (rank : 61) | NC score | 0.507476 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8IZF3, Q5T5B5, Q5T5B6, Q86SN9, Q8IXE6 | Gene names | GPR115, PGR18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 115 (G-protein coupled receptor PGR18). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 62) | NC score | 0.262379 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 63) | NC score | 0.248528 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
GPR56_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 64) | NC score | 0.531819 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y653, O95966, Q8NGB3, Q96HB4 | Gene names | GPR56, TM7LN4, TM7XN1 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 56 precursor (TM7XN1 protein). | |||||
|
GPR97_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 65) | NC score | 0.480966 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86Y34, Q6ZMF4, Q86SL9, Q8IZF1 | Gene names | GPR97, PGR26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor (G-protein coupled receptor PGR26). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 66) | NC score | 0.236450 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 67) | NC score | 0.484920 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP111_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 68) | NC score | 0.398940 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IZF7, Q86SL6, Q8NGU5, Q8TDT5 | Gene names | GPR111, PGR20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 111 (G-protein coupled receptor PGR20). | |||||
|
GP157_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 69) | NC score | 0.317861 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C206, Q8C224 | Gene names | Gpr157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GP128_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 70) | NC score | 0.518749 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BM96, Q80T42 | Gene names | Gpr128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
GP157_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 71) | NC score | 0.344580 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5UAW9, Q8WWB8, Q9HA73 | Gene names | GPR157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GPR97_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 72) | NC score | 0.484336 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
GPR98_MOUSE
|
||||||
| θ value | 0.62314 (rank : 73) | NC score | 0.311458 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.326878 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
CSMD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.009313 (rank : 101) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
O13J1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | -0.004514 (rank : 106) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NGT2, Q96R40 | Gene names | OR13J1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 13J1. | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.005779 (rank : 103) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
OL140_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | -0.004113 (rank : 104) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60878, Q9R0K1 | Gene names | Olfr140, Mor235-1, Olfr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 140 (Olfactory receptor 4A) (Olfactory receptor 235-1) (Odorant receptor A16). | |||||
|
OR4P4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | -0.004438 (rank : 105) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NGL7 | Gene names | OR4P4 | |||
|
Domain Architecture |
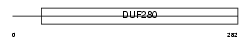
|
|||||
| Description | Olfactory receptor 4P4. | |||||
|
TPST1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.009480 (rank : 99) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60507, Q6FGM7 | Gene names | TPST1 | |||
|
Domain Architecture |
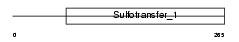
|
|||||
| Description | Protein-tyrosine sulfotransferase 1 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-1) (TPST-1). | |||||
|
TPST1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.009475 (rank : 100) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70281 | Gene names | Tpst1 | |||
|
Domain Architecture |
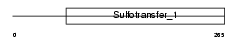
|
|||||
| Description | Protein-tyrosine sulfotransferase 1 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-1) (TPST-1). | |||||
|
TPST2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.009231 (rank : 102) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60704, Q6FI98, Q9H0V4 | Gene names | TPST2 | |||
|
Domain Architecture |
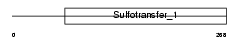
|
|||||
| Description | Protein-tyrosine sulfotransferase 2 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-2) (TPST-2). | |||||
|
FBLN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.051309 (rank : 96) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
FBLN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.050720 (rank : 97) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVJ9 | Gene names | Efemp2, Fbln4, Mbp1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (Mutant p53-binding protein 1). | |||||
|
GP114_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.467878 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8IZF4, Q6ZMH7, Q6ZML4, Q86SL8, Q8IZ14 | Gene names | GPR114, PGR27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 114 precursor (G-protein coupled receptor PGR27). | |||||
|
GP123_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.201535 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
GP124_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.138564 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96PE1, Q8N3R1, Q8TEM3, Q96KB2, Q9P1Z7, Q9UFY4 | Gene names | GPR124, KIAA1531, TEM5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
GP124_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.157796 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
GP125_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.150605 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IWK6, Q6UXK9, Q86SQ5, Q8TC55 | Gene names | GPR125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
GP125_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.139141 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TT36, Q6PE67, Q8VE71 | Gene names | Gpr125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
GPR56_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.495169 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8K209, Q3UR17, Q9QZT2 | Gene names | Gpr56, Cyt28 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 56 precursor (Serpentine receptor cyt28). | |||||
|
HMCN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.053873 (rank : 91) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053669 (rank : 92) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NELL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.053204 (rank : 94) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.052966 (rank : 95) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
PROP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.064892 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |
|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
PROP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.069707 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SPON1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.058709 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
SPON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.058673 (rank : 89) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
SUSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.066842 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
TSP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.058912 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.060700 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.059604 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
TSP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.058414 (rank : 90) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
UROM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053429 (rank : 93) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
UROM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050478 (rank : 98) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
CRFR1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P34998, Q13008, Q9UK64 | Gene names | CRHR1, CRFR, CRFR1, CRHR | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 1 precursor (CRF-R) (CRF1) (Corticotropin-releasing hormone receptor 1) (CRH-R 1). | |||||
|
CRFR1_MOUSE
|
||||||
| NC score | 0.998116 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P35347 | Gene names | Crhr1, Crhr | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 1 precursor (CRF-R) (CRF1) (Corticotropin-releasing hormone receptor 1) (CRH-R 1). | |||||
|
CRFR2_HUMAN
|
||||||
| NC score | 0.994166 (rank : 3) | θ value | 1.56915e-153 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q13324, O43461, Q99431 | Gene names | CRHR2, CRF2R, CRH2R | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 2 precursor (CRF-R 2) (CRF2) (Corticotropin-releasing hormone receptor 2) (CRH-R 2). | |||||
|
CRFR2_MOUSE
|
||||||
| NC score | 0.993639 (rank : 4) | θ value | 6.59259e-152 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q60748, Q60783, Q60808 | Gene names | Crhr2, Crf2r | |||
|
Domain Architecture |

|
|||||
| Description | Corticotropin-releasing factor receptor 2 precursor (CRF-R 2) (CRF2) (Corticotropin-releasing hormone receptor 2) (CRH-R 2) (CRF-RB) (CRH- R2). | |||||
|
PTHR1_HUMAN
|
||||||
| NC score | 0.940506 (rank : 5) | θ value | 2.75849e-49 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q03431 | Gene names | PTHR1, PTHR | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor (PTH/PTHr receptor) (PTH/PTHrP type I receptor). | |||||
|
PTHR1_MOUSE
|
||||||
| NC score | 0.935681 (rank : 6) | θ value | 7.51208e-47 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P41593, Q62119 | Gene names | Pthr1, Pthr | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone/parathyroid hormone-related peptide receptor precursor (PTH/PTHr receptor) (PTH/PTHrP type I receptor). | |||||
|
VIPR1_MOUSE
|
||||||
| NC score | 0.934966 (rank : 7) | θ value | 3.48949e-44 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P97751, Q9JI40, Q9R1T8 | Gene names | Vipr1 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 1 precursor (VIP-R-1) (VPAC1) (Pituitary adenylate cyclase-activating polypeptide type II receptor) (PACAP type II receptor) (PACAP-R-2). | |||||
|
CALCR_MOUSE
|
||||||
| NC score | 0.932638 (rank : 8) | θ value | 7.51208e-47 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q60755 | Gene names | Calcr | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin receptor precursor (CT-R). | |||||
|
CALCR_HUMAN
|
||||||
| NC score | 0.928724 (rank : 9) | θ value | 7.51208e-47 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P30988, O14585, Q13941, Q6DJU8, Q6T712 | Gene names | CALCR | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin receptor precursor (CT-R). | |||||
|
VIPR1_HUMAN
|
||||||
| NC score | 0.924782 (rank : 10) | θ value | 4.88048e-38 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P32241, Q15871 | Gene names | VIPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 1 precursor (VIP-R-1) (Pituitary adenylate cyclase-activating polypeptide type II receptor) (PACAP type II receptor) (PACAP-R-2). | |||||
|
VIPR2_MOUSE
|
||||||
| NC score | 0.923629 (rank : 11) | θ value | 8.89434e-40 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P41588, P97750 | Gene names | Vipr2 | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 2 precursor (VIP-R-2) (Pituitary adenylate cyclase-activating polypeptide type III receptor) (PACAP type III receptor) (PACAP-R-3). | |||||
|
VIPR2_HUMAN
|
||||||
| NC score | 0.922420 (rank : 12) | θ value | 5.76516e-39 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P41587, Q13053, Q15870 | Gene names | VIPR2, VIP2R | |||
|
Domain Architecture |

|
|||||
| Description | Vasoactive intestinal polypeptide receptor 2 precursor (VIP-R-2) (Pituitary adenylate cyclase-activating polypeptide type III receptor) (PACAP type III receptor) (PACAP-R-3) (Helodermin-preferring VIP receptor). | |||||
|
PACR_MOUSE
|
||||||
| NC score | 0.921725 (rank : 13) | θ value | 1.73584e-35 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P70205 | Gene names | Adcyap1r1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary adenylate cyclase-activating polypeptide type I receptor precursor (PACAP type I receptor) (PACAP-R-1). | |||||
|
PACR_HUMAN
|
||||||
| NC score | 0.921387 (rank : 14) | θ value | 1.12514e-34 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P41586 | Gene names | ADCYAP1R1 | |||
|
Domain Architecture |

|
|||||
| Description | Pituitary adenylate cyclase-activating polypeptide type I receptor precursor (PACAP type I receptor) (PACAP-R-1). | |||||
|
GHRHR_HUMAN
|
||||||
| NC score | 0.921047 (rank : 15) | θ value | 8.89434e-40 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q02643, Q99863 | Gene names | GHRHR | |||
|
Domain Architecture |

|
|||||
| Description | Growth hormone-releasing hormone receptor precursor (GHRH receptor) (GRF receptor) (GRFR). | |||||
|
GLP1R_MOUSE
|
||||||
| NC score | 0.920625 (rank : 16) | θ value | 7.27602e-42 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O35659 | Gene names | Glp1r | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 1 receptor precursor (GLP-1 receptor) (GLP-1-R) (GLP-1R). | |||||
|
SCTR_HUMAN
|
||||||
| NC score | 0.918317 (rank : 17) | θ value | 4.88048e-38 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P47872, Q12961, Q13213 | Gene names | SCTR | |||
|
Domain Architecture |

|
|||||
| Description | Secretin receptor precursor (SCT-R). | |||||
|
PTHR2_HUMAN
|
||||||
| NC score | 0.918010 (rank : 18) | θ value | 2.11701e-41 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P49190, Q8N429 | Gene names | PTHR2 | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone receptor precursor (PTH2 receptor). | |||||
|
GLR_HUMAN
|
||||||
| NC score | 0.917361 (rank : 19) | θ value | 3.37985e-39 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P47871 | Gene names | GCGR | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon receptor precursor (GL-R). | |||||
|
GLP1R_HUMAN
|
||||||
| NC score | 0.915731 (rank : 20) | θ value | 1.05066e-40 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P43220, Q99669 | Gene names | GLP1R | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 1 receptor precursor (GLP-1 receptor) (GLP-1-R) (GLP-1R). | |||||
|
PTHR2_MOUSE
|
||||||
| NC score | 0.915309 (rank : 21) | θ value | 9.50281e-42 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q91V95 | Gene names | Pthr2 | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone receptor precursor (PTH2 receptor). | |||||
|
GLR_MOUSE
|
||||||
| NC score | 0.912999 (rank : 22) | θ value | 2.05049e-36 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q61606, Q63960 | Gene names | Gcgr | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon receptor precursor (GL-R). | |||||
|
SCTR_MOUSE
|
||||||
| NC score | 0.912664 (rank : 23) | θ value | 4.72714e-33 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q5FWI2, Q80T66 | Gene names | Sctr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Secretin receptor precursor (SCT-R). | |||||
|
CALRL_HUMAN
|
||||||
| NC score | 0.912372 (rank : 24) | θ value | 5.22648e-32 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q16602 | Gene names | CALCRL, CGRPR | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin gene-related peptide type 1 receptor precursor (CGRP type 1 receptor) (Calcitonin receptor-like receptor). | |||||
|
CALRL_MOUSE
|
||||||
| NC score | 0.910509 (rank : 25) | θ value | 2.59387e-31 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9R1W5, Q9QXH8, Q9WUP2 | Gene names | Calcrl, Crlr | |||
|
Domain Architecture |

|
|||||
| Description | Calcitonin gene-related peptide type 1 receptor precursor (CGRP type 1 receptor) (Calcitonin receptor-like receptor). | |||||
|
GHRHR_MOUSE
|
||||||
| NC score | 0.909072 (rank : 26) | θ value | 4.42448e-31 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P32082 | Gene names | Ghrhr, Grfr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth hormone-releasing hormone receptor precursor (GHRH receptor) (GRF receptor) (GRFR). | |||||
|
GLP2R_HUMAN
|
||||||
| NC score | 0.906727 (rank : 27) | θ value | 1.98606e-31 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O95838 | Gene names | GLP2R | |||
|
Domain Architecture |

|
|||||
| Description | Glucagon-like peptide 2 receptor precursor (GLP-2 receptor) (GLP-2-R) (GLP-2R). | |||||
|
GIPR_HUMAN
|
||||||
| NC score | 0.906646 (rank : 28) | θ value | 1.08979e-29 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P48546, Q14401, Q16400, Q9UPI1 | Gene names | GIPR | |||
|
Domain Architecture |

|
|||||
| Description | Gastric inhibitory polypeptide receptor precursor (GIP-R) (Glucose- dependent insulinotropic polypeptide receptor). | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.744577 (rank : 29) | θ value | 3.28887e-18 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
EMR4_MOUSE
|
||||||
| NC score | 0.658068 (rank : 30) | θ value | 2.35696e-16 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q91ZE5, Q3U1I7, Q8BYX0, Q8VIM3 | Gene names | Emr4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 4 precursor (EGF-like module-containing mucin-like receptor EMR4) (F4/80-like-receptor) (Seven-span membrane protein FIRE). | |||||
|
GP144_HUMAN
|
||||||
| NC score | 0.645069 (rank : 31) | θ value | 2.36244e-08 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
EMR3_HUMAN
|
||||||
| NC score | 0.632767 (rank : 32) | θ value | 1.99529e-15 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9BY15 | Gene names | EMR3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 3 precursor (EGF-like module-containing mucin-like receptor EMR3). | |||||
|
GPR64_HUMAN
|
||||||
| NC score | 0.611011 (rank : 33) | θ value | 1.33837e-11 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8IZP9, O00406, Q8IWT2, Q8IZE4, Q8IZE5, Q8IZE6, Q8IZE7, Q8IZP3, Q8IZP4 | Gene names | GPR64, HE6, TM7LN2 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (He6 receptor). | |||||
|
GPR64_MOUSE
|
||||||
| NC score | 0.605862 (rank : 34) | θ value | 7.84624e-12 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8CJ12, Q8BL10, Q8CJ08, Q8CJ09, Q8CJ10 | Gene names | Gpr64, Me6 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 64 precursor (Epididymis-specific protein 6) (Me6 receptor). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.583323 (rank : 35) | θ value | 1.53129e-07 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
GP113_HUMAN
|
||||||
| NC score | 0.575713 (rank : 36) | θ value | 5.81887e-07 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8IZF5, Q6UXT7, Q6UXX3, Q86SL7, Q8IXD8, Q8TDT3 | Gene names | GPR113, PGR23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 113 precursor (G-protein coupled receptor PGR23). | |||||
|
LPHN3_HUMAN
|
||||||
| NC score | 0.574324 (rank : 37) | θ value | 1.02475e-11 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9HAR2, O94867, Q9NWK5 | Gene names | LPHN3, KIAA0768, LEC3 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-3 precursor (Calcium-independent alpha-latrotoxin receptor 3) (Lectomedin-3). | |||||
|
GP128_HUMAN
|
||||||
| NC score | 0.564211 (rank : 38) | θ value | 1.25267e-09 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q96K78, Q86SQ2 | Gene names | GPR128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
ELTD1_HUMAN
|
||||||
| NC score | 0.560318 (rank : 39) | θ value | 1.53129e-07 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9HBW9 | Gene names | ELTD1, ETL | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin and seven transmembrane domain-containing protein 1 precursor (EGF-TM7-latrophilin-related protein) (ETL protein). | |||||
|
LPHN2_HUMAN
|
||||||
| NC score | 0.549258 (rank : 40) | θ value | 2.61198e-07 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | O95490, O94882, Q5VX76, Q9UKY5, Q9UKY6 | Gene names | LPHN2, KIAA0786, LEC1, LPHH1 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-2 precursor (Calcium-independent alpha-latrotoxin receptor 2) (Latrophilin homolog 1) (Lectomedin-1). | |||||
|
GPR56_HUMAN
|
||||||
| NC score | 0.531819 (rank : 41) | θ value | 1.87187e-05 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y653, O95966, Q8NGB3, Q96HB4 | Gene names | GPR56, TM7LN4, TM7XN1 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 56 precursor (TM7XN1 protein). | |||||
|
GP128_MOUSE
|
||||||
| NC score | 0.518749 (rank : 42) | θ value | 0.00298849 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8BM96, Q80T42 | Gene names | Gpr128 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 128 precursor. | |||||
|
CD97_MOUSE
|
||||||
| NC score | 0.510079 (rank : 43) | θ value | 1.133e-10 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9Z0M6, Q923A1, Q9CVI5, Q9JLQ8 | Gene names | Cd97 | |||
|
Domain Architecture |

|
|||||
| Description | CD97 antigen precursor. | |||||
|
GP115_HUMAN
|
||||||
| NC score | 0.507476 (rank : 44) | θ value | 2.88788e-06 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8IZF3, Q5T5B5, Q5T5B6, Q86SN9, Q8IXE6 | Gene names | GPR115, PGR18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 115 (G-protein coupled receptor PGR18). | |||||
|
GP115_MOUSE
|
||||||
| NC score | 0.503169 (rank : 45) | θ value | 4.76016e-09 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9D2L6 | Gene names | Gpr115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 115 precursor. | |||||
|
ELTD1_MOUSE
|
||||||
| NC score | 0.496811 (rank : 46) | θ value | 3.41135e-07 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q923X1, Q91W44 | Gene names | Eltd1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin seven transmembrane domain-containing protein 1 precursor. | |||||
|
GPR56_MOUSE
|
||||||
| NC score | 0.495169 (rank : 47) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8K209, Q3UR17, Q9QZT2 | Gene names | Gpr56, Cyt28 | |||
|
Domain Architecture |

|
|||||
| Description | G-protein coupled receptor 56 precursor (Serpentine receptor cyt28). | |||||
|
GP110_HUMAN
|
||||||
| NC score | 0.494409 (rank : 48) | θ value | 8.11959e-09 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q5T601, Q5KU15, Q5T5Z9, Q86SM1, Q8IXE3, Q8IZF8, Q96DQ1 | Gene names | GPR110, PGR19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 110 precursor (G-protein coupled receptor PGR19) (G-protein coupled receptor KPG_012). | |||||
|
GP110_MOUSE
|
||||||
| NC score | 0.492013 (rank : 49) | θ value | 1.80886e-08 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VEC3, Q8BMV8 | Gene names | Gpr110 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 110 precursor. | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.484920 (rank : 50) | θ value | 0.000158464 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GPR97_MOUSE
|
||||||
| NC score | 0.484336 (rank : 51) | θ value | 0.00665767 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8R0T6 | Gene names | Gpr97 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor. | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.483566 (rank : 52) | θ value | 5.81887e-07 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GPR97_HUMAN
|
||||||
| NC score | 0.480966 (rank : 53) | θ value | 0.000121331 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q86Y34, Q6ZMF4, Q86SL9, Q8IZF1 | Gene names | GPR97, PGR26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 97 precursor (G-protein coupled receptor PGR26). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.476604 (rank : 54) | θ value | 1.2105e-12 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAI1_MOUSE
|
||||||
| NC score | 0.475824 (rank : 55) | θ value | 3.52202e-12 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 340 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q3UHD1, Q3UH36, Q8CGM0 | Gene names | Bai1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
CD97_HUMAN
|
||||||
| NC score | 0.467976 (rank : 56) | θ value | 2.52405e-10 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | P48960, O00718, O76101, Q8NG72, Q8TBQ7 | Gene names | CD97 | |||
|
Domain Architecture |

|
|||||
| Description | CD97 antigen precursor (Leukocyte antigen CD97). | |||||
|
GP114_HUMAN
|
||||||
| NC score | 0.467878 (rank : 57) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8IZF4, Q6ZMH7, Q6ZML4, Q86SL8, Q8IZ14 | Gene names | GPR114, PGR27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 114 precursor (G-protein coupled receptor PGR27). | |||||
|
GP116_HUMAN
|
||||||
| NC score | 0.467232 (rank : 58) | θ value | 2.79066e-09 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8IZF2, O94858, Q6RGN2, Q86SP0, Q9Y3Z2 | Gene names | GPR116, KIAA0758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 116 precursor. | |||||
|
EMR1_MOUSE
|
||||||
| NC score | 0.466380 (rank : 59) | θ value | 1.133e-10 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q61549 | Gene names | Emr1, Gpf480 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor) (Cell surface glycoprotein F4/80). | |||||
|
BAI2_MOUSE
|
||||||
| NC score | 0.453845 (rank : 60) | θ value | 1.38499e-08 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8CGM1, Q3TYC8, Q3UN11, Q3UNE2, Q6PGN0 | Gene names | Bai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI3_HUMAN
|
||||||
| NC score | 0.452948 (rank : 61) | θ value | 1.133e-10 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O60242, O60297 | Gene names | BAI3, KIAA0550 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
BAI2_HUMAN
|
||||||
| NC score | 0.452567 (rank : 62) | θ value | 1.80886e-08 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60241, Q5T6K0, Q8NGW8 | Gene names | BAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 2 precursor. | |||||
|
BAI3_MOUSE
|
||||||
| NC score | 0.451898 (rank : 63) | θ value | 1.133e-10 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q80ZF8 | Gene names | Bai3 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 3 precursor. | |||||
|
EMR1_HUMAN
|
||||||
| NC score | 0.451473 (rank : 64) | θ value | 7.84624e-12 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q14246, Q8NGA7 | Gene names | EMR1, TM7LN3 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 1 precursor (Cell surface glycoprotein EMR1) (EMR1 hormone receptor). | |||||
|
EMR2_HUMAN
|
||||||
| NC score | 0.444824 (rank : 65) | θ value | 1.74796e-11 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9UHX3, Q8NG96, Q9Y4B1 | Gene names | EMR2 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 2 precursor (EGF-like module EMR2) (CD312 antigen). | |||||
|
GP111_HUMAN
|
||||||
| NC score | 0.398940 (rank : 66) | θ value | 0.00134147 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8IZF7, Q86SL6, Q8NGU5, Q8TDT5 | Gene names | GPR111, PGR20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 111 (G-protein coupled receptor PGR20). | |||||
|
GP157_HUMAN
|
||||||
| NC score | 0.344580 (rank : 67) | θ value | 0.00298849 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q5UAW9, Q8WWB8, Q9HA73 | Gene names | GPR157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.326878 (rank : 68) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
GP157_MOUSE
|
||||||
| NC score | 0.317861 (rank : 69) | θ value | 0.00134147 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8C206, Q8C224 | Gene names | Gpr157 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 157. | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.311458 (rank : 70) | θ value | 0.62314 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.284330 (rank : 71) | θ value | 4.45536e-07 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.276569 (rank : 72) | θ value | 5.81887e-07 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.262379 (rank : 73) | θ value | 3.77169e-06 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.261686 (rank : 74) | θ value | 9.92553e-07 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.248528 (rank : 75) | θ value | 1.43324e-05 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.236450 (rank : 76) | θ value | 0.000158464 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
GP123_HUMAN
|
||||||
| NC score | 0.201535 (rank : 77) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q86SQ6, Q86SN7, Q96JJ9 | Gene names | GPR123, KIAA1828 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 123. | |||||
|
GP124_MOUSE
|
||||||
| NC score | 0.157796 (rank : 78) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q91ZV8 | Gene names | Gpr124, Tem5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
GP125_HUMAN
|
||||||
| NC score | 0.150605 (rank : 79) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8IWK6, Q6UXK9, Q86SQ5, Q8TC55 | Gene names | GPR125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
GP125_MOUSE
|
||||||
| NC score | 0.139141 (rank : 80) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q7TT36, Q6PE67, Q8VE71 | Gene names | Gpr125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 125 precursor. | |||||
|
GP124_HUMAN
|
||||||
| NC score | 0.138564 (rank : 81) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 304 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96PE1, Q8N3R1, Q8TEM3, Q96KB2, Q9P1Z7, Q9UFY4 | Gene names | GPR124, KIAA1531, TEM5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 124 precursor (Tumor endothelial marker 5). | |||||
|
PROP_MOUSE
|
||||||
| NC score | 0.069707 (rank : 82) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11680, Q3TB98, Q3U779 | Gene names | Cfp, Pfc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Properdin precursor (Factor P). | |||||
|
SUSD1_HUMAN
|
||||||
| NC score | 0.066842 (rank : 83) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UWL2, Q5T8V7, Q6P9G7, Q8WU83, Q9H6V2, Q9NTA7 | Gene names | SUSD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sushi domain-containing protein 1 precursor. | |||||
|
PROP_HUMAN
|
||||||
| NC score | 0.064892 (rank : 84) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P27918, O15134, O15135, O15136, O75826 | Gene names | CFP, PFC | |||
|
Domain Architecture |

|
|||||
| Description | Properdin precursor (Factor P). | |||||
|
TSP1_MOUSE
|
||||||
| NC score | 0.060700 (rank : 85) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP2_HUMAN
|
||||||
| NC score | 0.059604 (rank : 86) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P35442 | Gene names | THBS2, TSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
TSP1_HUMAN
|
||||||
| NC score | 0.058912 (rank : 87) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
SPON1_HUMAN
|
||||||
| NC score | 0.058709 (rank : 88) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9HCB6, O94862, Q8NCD7, Q8WUR5 | Gene names | SPON1, KIAA0762, VSGP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin) (Vascular smooth muscle cell growth- promoting factor). | |||||
|
SPON1_MOUSE
|
||||||
| NC score | 0.058673 (rank : 89) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCC9, Q8K2Q8 | Gene names | Spon1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spondin-1 precursor (F-spondin). | |||||
|
TSP2_MOUSE
|
||||||
| NC score | 0.058414 (rank : 90) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
HMCN1_HUMAN
|
||||||
| NC score | 0.053873 (rank : 91) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 983 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96RW7, Q5TYR7, Q96DN3, Q96DN8, Q96SC3 | Gene names | HMCN1, FIBL6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hemicentin-1 precursor (Fibulin-6) (FIBL-6). | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.053669 (rank : 92) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
UROM_HUMAN
|
||||||
| NC score | 0.053429 (rank : 93) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07911, Q540J6, Q6ZS84, Q8IYG0 | Gene names | UMOD | |||
|
Domain Architecture |

|
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.053204 (rank : 94) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.052966 (rank : 95) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
FBLN4_HUMAN
|
||||||
| NC score | 0.051309 (rank : 96) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95967, O75967 | Gene names | EFEMP2, FBLN4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (UPH1 protein). | |||||
|
FBLN4_MOUSE
|
||||||
| NC score | 0.050720 (rank : 97) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WVJ9 | Gene names | Efemp2, Fbln4, Mbp1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-containing fibulin-like extracellular matrix protein 2 precursor (Fibulin-4) (FIBL-4) (Mutant p53-binding protein 1). | |||||
|
UROM_MOUSE
|
||||||
| NC score | 0.050478 (rank : 98) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q91X17, Q3TN64, Q3TP60, Q62285 | Gene names | Umod | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uromodulin precursor (Tamm-Horsfall urinary glycoprotein) (THP). | |||||
|
TPST1_HUMAN
|
||||||
| NC score | 0.009480 (rank : 99) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60507, Q6FGM7 | Gene names | TPST1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-tyrosine sulfotransferase 1 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-1) (TPST-1). | |||||
|
TPST1_MOUSE
|
||||||
| NC score | 0.009475 (rank : 100) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70281 | Gene names | Tpst1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-tyrosine sulfotransferase 1 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-1) (TPST-1). | |||||
|
CSMD3_HUMAN
|
||||||
| NC score | 0.009313 (rank : 101) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7Z407, Q96PZ3 | Gene names | CSMD3, KIAA1894 | |||
|
Domain Architecture |

|
|||||
| Description | CUB and sushi domain-containing protein 3 precursor (CUB and sushi multiple domains protein 3). | |||||
|
TPST2_HUMAN
|
||||||
| NC score | 0.009231 (rank : 102) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60704, Q6FI98, Q9H0V4 | Gene names | TPST2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-tyrosine sulfotransferase 2 (EC 2.8.2.20) (Tyrosylprotein sulfotransferase-2) (TPST-2). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.005779 (rank : 103) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
OL140_MOUSE
|
||||||
| NC score | -0.004113 (rank : 104) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60878, Q9R0K1 | Gene names | Olfr140, Mor235-1, Olfr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Olfactory receptor 140 (Olfactory receptor 4A) (Olfactory receptor 235-1) (Odorant receptor A16). | |||||
|
OR4P4_HUMAN
|
||||||
| NC score | -0.004438 (rank : 105) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NGL7 | Gene names | OR4P4 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 4P4. | |||||
|
O13J1_HUMAN
|
||||||
| NC score | -0.004514 (rank : 106) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NGT2, Q96R40 | Gene names | OR13J1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 13J1. | |||||