Please be patient as the page loads
|
CABL2_HUMAN
|
||||||
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CABL2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CABL2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.963682 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CABL1_HUMAN
|
||||||
| θ value | 1.21265e-121 (rank : 3) | NC score | 0.944970 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL1_MOUSE
|
||||||
| θ value | 2.22025e-107 (rank : 4) | NC score | 0.944199 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 5) | NC score | 0.055598 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 6) | NC score | 0.074239 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 7) | NC score | 0.063781 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.056926 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.065192 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
JPH2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.057798 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.109115 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.110024 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 13) | NC score | 0.110416 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.109200 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.109173 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 16) | NC score | 0.108804 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 0.125558 (rank : 17) | NC score | 0.058969 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
GAK10_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.109416 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.107970 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
RGS14_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.029118 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.043949 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.044054 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.020703 (rank : 92) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.049556 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.046866 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
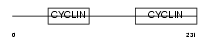
CCNC_MOUSE
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.071493 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.007669 (rank : 111) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
IL3RB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.054376 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
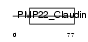
CLD23_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.033330 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-23. | |||||
|
FOXO4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.021289 (rank : 90) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
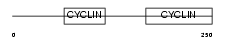
CCNC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.073656 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-C. | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.017497 (rank : 95) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
ZN358_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.003218 (rank : 117) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
EFS_MOUSE
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.030329 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.050826 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.051199 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SAM68_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.034598 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.105376 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.104731 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
CAC1D_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.008432 (rank : 110) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.062254 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
KCNN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.014248 (rank : 102) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.030910 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
AATK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.011046 (rank : 107) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.013980 (rank : 103) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
GAK7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.100838 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.026248 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.030063 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.031238 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
RTKN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.024929 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BST9, Q8WVN1, Q96PT6, Q9HB05 | Gene names | RTKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
SAM68_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.030598 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
T22D4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.027970 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.015161 (rank : 100) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
XKR7_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.016150 (rank : 97) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
AIRE_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.025090 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.037172 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
EGR4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.004153 (rank : 115) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.013602 (rank : 105) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.027303 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.021430 (rank : 89) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ABRA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.019717 (rank : 93) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.028546 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.021198 (rank : 91) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.031124 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
COG8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.023251 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
DUS8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.013648 (rank : 104) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
FHOD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.019162 (rank : 94) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
JIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.023695 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
MYCB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.014886 (rank : 101) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.049316 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.012044 (rank : 106) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
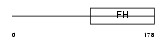
FOXO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.016017 (rank : 99) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |
|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
IF4B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.016115 (rank : 98) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
PIAS3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.008622 (rank : 109) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.016708 (rank : 96) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
CD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.037374 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.002485 (rank : 118) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.001937 (rank : 119) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.031896 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
TLE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.005138 (rank : 113) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
WNK4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.006016 (rank : 112) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
WNK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.004483 (rank : 114) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
WWC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.010781 (rank : 108) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.003584 (rank : 116) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052429 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.056169 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051135 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050435 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
GAK13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.082618 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.085553 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.092042 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.092580 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.098644 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.088882 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.092466 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
IGEB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.059326 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P03975 | Gene names | Iap | |||
|
Domain Architecture |

|
|||||
| Description | IgE-binding protein. | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.071734 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.084158 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.072291 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.071362 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.057679 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.063051 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.050790 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.065470 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.082439 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
PRR13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.064174 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.055303 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.052117 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.057638 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.062328 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.054948 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.066947 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.068329 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.057264 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.060152 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050517 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
WASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051381 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.060455 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.062360 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
CABL2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9BTV7, Q5JWL0, Q9BYK0 | Gene names | CABLES2, C20orf150 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CABL2_MOUSE
|
||||||
| NC score | 0.963682 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8K3M5 | Gene names | Cables2 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 and ABL1 enzyme substrate 2 (Interactor with CDK3 2) (Ik3-2). | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.944970 (rank : 3) | θ value | 1.21265e-121 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
CABL1_MOUSE
|
||||||
| NC score | 0.944199 (rank : 4) | θ value | 2.22025e-107 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESJ1, Q9EPR8 | Gene names | Cables1, Cables | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.110416 (rank : 5) | θ value | 0.125558 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.110024 (rank : 6) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK10_HUMAN
|
||||||
| NC score | 0.109416 (rank : 7) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P87889, P10263, P10264, Q69385, Q9UKH6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q33.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K10 Gag protein) (HERV-K107 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.109200 (rank : 8) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.109173 (rank : 9) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.109115 (rank : 10) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.108804 (rank : 11) | θ value | 0.125558 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.107970 (rank : 12) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.105376 (rank : 13) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK8_HUMAN
|
||||||
| NC score | 0.104731 (rank : 14) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9HDB9 | Gene names | ERVK5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q12.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(II) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK7_HUMAN
|
||||||
| NC score | 0.100838 (rank : 15) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PI4, Q9QC08 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q23.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K18 Gag protein) (HERV-K110 Gag protein) (HERV-K(C1a) Gag protein) [Contains: Matrix protein]. | |||||
|
GAK18_HUMAN
|
||||||
| NC score | 0.098644 (rank : 16) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P62690 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.23 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK17_HUMAN
|
||||||
| NC score | 0.092580 (rank : 17) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62689 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_11q22.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein]. | |||||
|
GAK9_HUMAN
|
||||||
| NC score | 0.092466 (rank : 18) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UKH8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K104 Gag protein) [Contains: Matrix protein]. | |||||
|
GAK15_HUMAN
|
||||||
| NC score | 0.092042 (rank : 19) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62688 | Gene names | ERVK4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_3q21.2 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(I) Gag protein). | |||||
|
GAK19_HUMAN
|
||||||
| NC score | 0.088882 (rank : 20) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62691 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_4p16.1 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
GAK14_HUMAN
|
||||||
| NC score | 0.085553 (rank : 21) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62687 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_16p3.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.084158 (rank : 22) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
GAK13_HUMAN
|
||||||
| NC score | 0.082618 (rank : 23) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P62686 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1p13.3 provirus ancestral Gag polyprotein (Gag polyprotein). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.082439 (rank : 24) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.074239 (rank : 25) | θ value | 0.00134147 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.073656 (rank : 26) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.072291 (rank : 27) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.071734 (rank : 28) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.071493 (rank : 29) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.071362 (rank : 30) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.068329 (rank : 31) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.066947 (rank : 32) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.065470 (rank : 33) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.065192 (rank : 34) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PRR13_MOUSE
|
||||||
| NC score | 0.064174 (rank : 35) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CQJ5, Q3U3U4 | Gene names | Prr13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.063781 (rank : 36) | θ value | 0.00390308 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.063051 (rank : 37) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.062360 (rank : 38) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.062328 (rank : 39) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.062254 (rank : 40) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.060455 (rank : 41) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.060152 (rank : 42) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
IGEB_MOUSE
|
||||||
| NC score | 0.059326 (rank : 43) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P03975 | Gene names | Iap | |||
|
Domain Architecture |

|
|||||
| Description | IgE-binding protein. | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.058969 (rank : 44) | θ value | 0.125558 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
JPH2_HUMAN
|
||||||
| NC score | 0.057798 (rank : 45) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BR39, O95913, Q5JY74, Q9UJN4 | Gene names | JPH2, JP2 | |||
|
Domain Architecture |

|
|||||
| Description | Junctophilin-2 (Junctophilin type 2) (JP-2). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.057679 (rank : 46) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.057638 (rank : 47) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.057264 (rank : 48) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.056926 (rank : 49) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.056169 (rank : 50) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.055598 (rank : 51) | θ value | 0.00102713 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.055303 (rank : 52) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.054948 (rank : 53) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
IL3RB_MOUSE
|
||||||
| NC score | 0.054376 (rank : 54) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 355 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26955 | Gene names | Csf2rb, Aic2b, Csf2rb1, Il3rb1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytokine receptor common beta chain precursor (GM-CSF/IL-3/IL-5 receptor common beta-chain) (CD131 antigen). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.052429 (rank : 55) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.052117 (rank : 56) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
WASP_MOUSE
|
||||||
| NC score | 0.051381 (rank : 57) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70315 | Gene names | Was, Wasp | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein homolog (WASp). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.051199 (rank : 58) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.051135 (rank : 59) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.050826 (rank : 60) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.050790 (rank : 61) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
WASP_HUMAN
|
||||||
| NC score | 0.050517 (rank : 62) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P42768, Q9BU11, Q9UNJ9 | Gene names | WAS, IMD2 | |||
|
Domain Architecture |

|
|||||
| Description | Wiskott-Aldrich syndrome protein (WASp). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.050435 (rank : 63) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.049556 (rank : 64) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.049316 (rank : 65) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.046866 (rank : 66) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.044054 (rank : 67) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.043949 (rank : 68) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
CD2_HUMAN
|
||||||
| NC score | 0.037374 (rank : 69) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P06729, Q96TE5 | Gene names | CD2 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface antigen CD2 precursor (T-cell surface antigen T11/Leu- 5) (LFA-2) (LFA-3 receptor) (Erythrocyte receptor) (Rosette receptor). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.037172 (rank : 70) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
SAM68_HUMAN
|
||||||
| NC score | 0.034598 (rank : 71) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07666, Q6PJX7, Q8NB97, Q99760 | Gene names | KHDRBS1, SAM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
CLD23_HUMAN
|
||||||
| NC score | 0.033330 (rank : 72) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96B33 | Gene names | CLDN23 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-23. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.031896 (rank : 73) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.031238 (rank : 74) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.031124 (rank : 75) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.030910 (rank : 76) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
SAM68_MOUSE
|
||||||
| NC score | 0.030598 (rank : 77) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60749, Q3U8T3, Q60735, Q99M33 | Gene names | Khdrbs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | KH domain-containing, RNA-binding, signal transduction-associated protein 1 (p21 Ras GTPase-activating protein-associated p62) (GAP- associated tyrosine phosphoprotein p62) (Src-associated in mitosis 68 kDa protein) (Sam68) (p68). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.030329 (rank : 78) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.030063 (rank : 79) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RGS14_HUMAN
|
||||||
| NC score | 0.029118 (rank : 80) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43566, O43565, Q8TD62 | Gene names | RGS14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.028546 (rank : 81) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
T22D4_HUMAN
|
||||||
| NC score | 0.027970 (rank : 82) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y3Q8 | Gene names | TSC22D4, TILZ2 | |||
|
Domain Architecture |

|
|||||
| Description | TSC22 domain family protein 4 (TSC22-related-inducible leucine zipper protein 2) (Tsc-22-like protein THG-1). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.027303 (rank : 83) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.026248 (rank : 84) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.025090 (rank : 85) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
RTKN_HUMAN
|
||||||
| NC score | 0.024929 (rank : 86) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BST9, Q8WVN1, Q96PT6, Q9HB05 | Gene names | RTKN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhotekin. | |||||
|
JIP1_MOUSE
|
||||||
| NC score | 0.023695 (rank : 87) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WVI9, O35145, Q925J8, Q9R1H9, Q9R1Z1, Q9WVI7, Q9WVI8 | Gene names | Mapk8ip1, Ib1, Jip1, Mapk8ip, Prkm8ip | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain-1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
COG8_MOUSE
|
||||||
| NC score | 0.023251 (rank : 88) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JJA2 | Gene names | Cog8 | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 8. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.021430 (rank : 89) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
FOXO4_HUMAN
|
||||||
| NC score | 0.021289 (rank : 90) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P98177, O43821, Q13720, Q3KPF1, Q8TDK9 | Gene names | MLLT7, AFX, AFX1, FOXO4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Forkhead box protein O4). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.021198 (rank : 91) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.020703 (rank : 92) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ABRA_HUMAN
|
||||||
| NC score | 0.019717 (rank : 93) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N0Z2 | Gene names | ABRA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding Rho-activating protein (Striated muscle activator of Rho-dependent signaling) (STARS). | |||||
|
FHOD1_HUMAN
|
||||||
| NC score | 0.019162 (rank : 94) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y613, Q59F76, Q6Y1F2, Q76MS8, Q8N521 | Gene names | FHOD1, FHOS | |||
|
Domain Architecture |

|
|||||
| Description | FH1/FH2 domain-containing protein (Formin homolog overexpressed in spleen) (FHOS) (Formin homology 2 domain-containing protein 1). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.017497 (rank : 95) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.016708 (rank : 96) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
XKR7_MOUSE
|
||||||
| NC score | 0.016150 (rank : 97) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5GH64 | Gene names | Xkr7, Xrg7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | XK-related protein 7. | |||||
|
IF4B_MOUSE
|
||||||
| NC score | 0.016115 (rank : 98) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGD9, Q3TD64 | Gene names | Eif4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
FOXO4_MOUSE
|
||||||
| NC score | 0.016017 (rank : 99) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WVH3 | Gene names | Mllt7, Afx, Afx1, Fkhr3, Foxo4 | |||
|
Domain Architecture |

|
|||||
| Description | Fork head domain transcription factor AFX1 (Afxh) (Forkhead box protein O4). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.015161 (rank : 100) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
MYCB2_HUMAN
|
||||||
| NC score | 0.014886 (rank : 101) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 390 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75592, Q5JSX8, Q5VZN6, Q6PIB6, Q9UQ11, Q9Y6E4 | Gene names | MYCBP2, KIAA0916, PAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
KCNN1_HUMAN
|
||||||
| NC score | 0.014248 (rank : 102) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92952 | Gene names | KCNN1, SK | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 1 (SK1). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.013980 (rank : 103) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
DUS8_MOUSE
|
||||||
| NC score | 0.013648 (rank : 104) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O09112 | Gene names | Dusp8, Nttp1 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Neuronal tyrosine threonine phosphatase 1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.013602 (rank : 105) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.012044 (rank : 106) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
AATK_HUMAN
|
||||||
| NC score | 0.011046 (rank : 107) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1243 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q6ZMQ8, O75136, Q6ZN31, Q86X28 | Gene names | AATK, KIAA0641 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis-associated tyrosine-protein kinase (EC 2.7.10.2) (AATYK) (Brain apoptosis-associated tyrosine kinase) (CDK5-binding protein) (p35-binding protein) (p35BP). | |||||
|
WWC3_HUMAN
|
||||||
| NC score | 0.010781 (rank : 108) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULE0, Q659C1, Q9BTQ1 | Gene names | WWC3, KIAA1280 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WWC family member 3. | |||||
|
PIAS3_MOUSE
|
||||||
| NC score | 0.008622 (rank : 109) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54714, Q80WF8, Q8R598 | Gene names | Pias3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein inhibitor of activated STAT protein 3. | |||||
|
CAC1D_HUMAN
|
||||||
| NC score | 0.008432 (rank : 110) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q01668, Q13916, Q13931 | Gene names | CACNA1D, CACH3, CACN4, CACNL1A2, CCHL1A2 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1D (Voltage- gated calcium channel subunit alpha Cav1.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 2). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | 0.007669 (rank : 111) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
WNK4_HUMAN
|
||||||
| NC score | 0.006016 (rank : 112) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1043 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96J92, Q8N8X3, Q8N8Z2, Q96DT8, Q9BYS5 | Gene names | WNK4, PRKWNK4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
TLE1_HUMAN
|
||||||
| NC score | 0.005138 (rank : 113) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q04724, Q5T3G4, Q969V9 | Gene names | TLE1 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 1 (ESG1) (E(Sp1) homolog). | |||||
|
WNK4_MOUSE
|
||||||
| NC score | 0.004483 (rank : 114) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
EGR4_HUMAN
|
||||||
| NC score | 0.004153 (rank : 115) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05215 | Gene names | EGR4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4) (AT133). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.003584 (rank : 116) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
ZN358_HUMAN
|
||||||
| NC score | 0.003218 (rank : 117) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NW07, Q9BTM7 | Gene names | ZNF358 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 358. | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.002485 (rank : 118) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.001937 (rank : 119) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||