Please be patient as the page loads
|
NLGN2_HUMAN
|
||||||
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NLGN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994971 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994882 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |
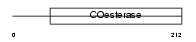
|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999592 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.994860 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |
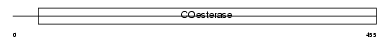
|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.994902 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGNX_HUMAN
|
||||||
| θ value | 0 (rank : 7) | NC score | 0.995063 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGNY_HUMAN
|
||||||
| θ value | 0 (rank : 8) | NC score | 0.995180 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
CHLE_MOUSE
|
||||||
| θ value | 2.47191e-82 (rank : 9) | NC score | 0.947873 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_HUMAN
|
||||||
| θ value | 3.56943e-81 (rank : 10) | NC score | 0.948154 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
ACES_MOUSE
|
||||||
| θ value | 6.08852e-81 (rank : 11) | NC score | 0.949893 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_HUMAN
|
||||||
| θ value | 3.0217e-80 (rank : 12) | NC score | 0.949148 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
EST1_HUMAN
|
||||||
| θ value | 4.08394e-77 (rank : 13) | NC score | 0.934776 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
EST2_HUMAN
|
||||||
| θ value | 4.08394e-77 (rank : 14) | NC score | 0.950852 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
CEL_MOUSE
|
||||||
| θ value | 1.45253e-74 (rank : 15) | NC score | 0.953374 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST22_MOUSE
|
||||||
| θ value | 7.20884e-74 (rank : 16) | NC score | 0.934475 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
CES3_MOUSE
|
||||||
| θ value | 6.10265e-73 (rank : 17) | NC score | 0.932833 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 3.95564e-72 (rank : 18) | NC score | 0.723685 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST31_MOUSE
|
||||||
| θ value | 3.34864e-71 (rank : 19) | NC score | 0.948628 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
EST1_MOUSE
|
||||||
| θ value | 6.31527e-70 (rank : 20) | NC score | 0.930267 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
ESTN_MOUSE
|
||||||
| θ value | 1.83746e-69 (rank : 21) | NC score | 0.932724 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
THYG_MOUSE
|
||||||
| θ value | 2.041e-52 (rank : 22) | NC score | 0.812449 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| θ value | 2.50074e-42 (rank : 23) | NC score | 0.793576 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 24) | NC score | 0.044921 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
DPP8_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 25) | NC score | 0.063738 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6V1X1, Q7Z4C8, Q7Z4D3, Q7Z4E1, Q8IWG7, Q8NEM5, Q96JX1, Q9HBM2, Q9HBM3, Q9HBM4, Q9HBM5, Q9NXF4 | Gene names | DPP8, DPRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 8 (EC 3.4.14.5) (Dipeptidyl peptidase VIII) (DP8) (Prolyl dipeptidase DPP8) (Dipeptidyl peptidase IV-related protein 1) (DPRP-1). | |||||
|
DPP8_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 26) | NC score | 0.061681 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YA7, Q9D4G6 | Gene names | Dpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 8 (EC 3.4.14.5) (Dipeptidyl peptidase VIII) (DP8). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 27) | NC score | 0.021521 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.044672 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
DPP9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.056368 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86TI2, Q6AI37, Q6UAL0, Q6ZMT2, Q6ZNJ5, Q8N2J7, Q8N3F5, Q8WXD8, Q96NT8, Q9BVR3 | Gene names | DPP9, DPRP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9) (Dipeptidyl peptidase IV-related protein 2) (DPRP-2). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.014794 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
DPP9_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.049900 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BVG4, Q6KAM9, Q8BWT9 | Gene names | Dpp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 0.365318 (rank : 32) | NC score | 0.038632 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.014888 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
BAG3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.015897 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
PLXB1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.012421 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.024800 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
TRPV4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.009702 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.014613 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
LAG3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.015950 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61790, Q0VBL2 | Gene names | Lag3 | |||
|
Domain Architecture |
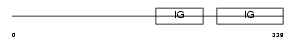
|
|||||
| Description | Lymphocyte activation gene 3 protein precursor (LAG-3) (CD223 antigen). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.021282 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
GP108_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.011794 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.005327 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | -0.000315 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | -0.000667 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ATP9B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.003277 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
EPN2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.007682 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.005897 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.010268 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.014195 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.002375 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.002429 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
PTPRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.002950 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
RKHD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.010374 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.005438 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
USH1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.003602 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
AAAD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.208575 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.117110 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
SMR3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.090712 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
NLGN2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8NFZ4, Q9P2I1 | Gene names | NLGN2, KIAA1366 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGN2_MOUSE
|
||||||
| NC score | 0.999592 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q69ZK9, Q5F288 | Gene names | Nlgn2, Kiaa1366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroligin-2 precursor. | |||||
|
NLGNY_HUMAN
|
||||||
| NC score | 0.995180 (rank : 3) | θ value | 0 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NFZ3, Q7Z3T5, Q9Y2F8 | Gene names | NLGN4Y, KIAA0951 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, Y-linked precursor (Neuroligin Y). | |||||
|
NLGNX_HUMAN
|
||||||
| NC score | 0.995063 (rank : 4) | θ value | 0 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N0W4, Q6UX10, Q9ULG0 | Gene names | NLGN4X, KIAA1260, NLGN4 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-4, X-linked precursor (Neuroligin X) (HNLX). | |||||
|
NLGN1_HUMAN
|
||||||
| NC score | 0.994971 (rank : 5) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8N2Q7, Q9UPT2 | Gene names | NLGN1, KIAA1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN3_MOUSE
|
||||||
| NC score | 0.994902 (rank : 6) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BYM5, Q8BXR4 | Gene names | Nlgn3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
NLGN1_MOUSE
|
||||||
| NC score | 0.994882 (rank : 7) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99K10 | Gene names | Nlgn1, Kiaa1070 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-1 precursor. | |||||
|
NLGN3_HUMAN
|
||||||
| NC score | 0.994860 (rank : 8) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZ94, Q8NCD0, Q9NZ95, Q9NZ96, Q9NZ97, Q9P248 | Gene names | NLGN3, KIAA1480, NL3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuroligin-3 precursor (Gliotactin homolog). | |||||
|
CEL_MOUSE
|
||||||
| NC score | 0.953374 (rank : 9) | θ value | 1.45253e-74 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64285 | Gene names | Cel, Lip1 | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
EST2_HUMAN
|
||||||
| NC score | 0.950852 (rank : 10) | θ value | 4.08394e-77 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00748, Q16859, Q5MAB8, Q7Z366, Q8IUP4, Q8TCP8 | Gene names | CES2, ICE | |||
|
Domain Architecture |

|
|||||
| Description | Carboxylesterase 2 precursor (EC 3.1.1.1) (CE-2) (hCE-2). | |||||
|
ACES_MOUSE
|
||||||
| NC score | 0.949893 (rank : 11) | θ value | 6.08852e-81 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P21836 | Gene names | Ache | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
ACES_HUMAN
|
||||||
| NC score | 0.949148 (rank : 12) | θ value | 3.0217e-80 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P22303, Q16169, Q86TM9, Q9BXP7 | Gene names | ACHE | |||
|
Domain Architecture |

|
|||||
| Description | Acetylcholinesterase precursor (EC 3.1.1.7) (AChE). | |||||
|
EST31_MOUSE
|
||||||
| NC score | 0.948628 (rank : 13) | θ value | 3.34864e-71 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q63880 | Gene names | Es31 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 31 precursor (EC 3.1.1.1) (ES-Male) (Esterase- 31). | |||||
|
CHLE_HUMAN
|
||||||
| NC score | 0.948154 (rank : 14) | θ value | 3.56943e-81 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P06276 | Gene names | BCHE, CHE1 | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
CHLE_MOUSE
|
||||||
| NC score | 0.947873 (rank : 15) | θ value | 2.47191e-82 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q03311 | Gene names | Bche | |||
|
Domain Architecture |

|
|||||
| Description | Cholinesterase precursor (EC 3.1.1.8) (Acylcholine acylhydrolase) (Choline esterase II) (Butyrylcholine esterase) (Pseudocholinesterase). | |||||
|
EST1_HUMAN
|
||||||
| NC score | 0.934776 (rank : 16) | θ value | 4.08394e-77 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P23141, Q00015, Q13657, Q14062, Q16737, Q16788, Q86UK2, Q96EE8, Q9UK77, Q9ULY2 | Gene names | CES1, CES2, SES1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ACAT) (Monocyte/macrophage serine esterase) (HMSE) (Serine esterase 1) (Brain carboxylesterase hBr1) (Triacylglycerol hydrolase) (TGH) (Egasyn). | |||||
|
EST22_MOUSE
|
||||||
| NC score | 0.934475 (rank : 17) | θ value | 7.20884e-74 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q64176 | Gene names | Es22 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase 22 precursor (EC 3.1.1.1) (Egasyn) (Esterase- 22) (Es-22). | |||||
|
CES3_MOUSE
|
||||||
| NC score | 0.932833 (rank : 18) | θ value | 6.10265e-73 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCT4, Q91ZV9, Q924V8, Q9ULY1 | Gene names | Ces3, Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Carboxylesterase 3 precursor (EC 3.1.1.1) (Triacylglycerol hydrolase) (TGH). | |||||
|
ESTN_MOUSE
|
||||||
| NC score | 0.932724 (rank : 19) | θ value | 1.83746e-69 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P23953, O54936, P11374, Q8K125 | Gene names | Es1 | |||
|
Domain Architecture |

|
|||||
| Description | Liver carboxylesterase N precursor (EC 3.1.1.1) (PES-N) (Lung surfactant convertase). | |||||
|
EST1_MOUSE
|
||||||
| NC score | 0.930267 (rank : 20) | θ value | 6.31527e-70 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VCC2, O55136 | Gene names | Ces1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Liver carboxylesterase 1 precursor (EC 3.1.1.1) (Acyl coenzyme A:cholesterol acyltransferase) (ES-x). | |||||
|
THYG_MOUSE
|
||||||
| NC score | 0.812449 (rank : 21) | θ value | 2.041e-52 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08710, O88590, Q9QWY7 | Gene names | Tg, Tgn | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
THYG_HUMAN
|
||||||
| NC score | 0.793576 (rank : 22) | θ value | 2.50074e-42 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P01266, O15274, O43899, Q15593, Q15948, Q9NYR1, Q9NYR2, Q9UMZ0, Q9UNY3 | Gene names | TG | |||
|
Domain Architecture |

|
|||||
| Description | Thyroglobulin precursor. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.723685 (rank : 23) | θ value | 3.95564e-72 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
AAAD_HUMAN
|
||||||
| NC score | 0.208575 (rank : 24) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P22760, Q8N1A9 | Gene names | AADAC, DAC | |||
|
Domain Architecture |

|
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
AAAD_MOUSE
|
||||||
| NC score | 0.117110 (rank : 25) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99PG0, Q8VCF2 | Gene names | Aadac, Aada | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arylacetamide deacetylase (EC 3.1.1.-) (AADAC). | |||||
|
SMR3B_HUMAN
|
||||||
| NC score | 0.090712 (rank : 26) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02814, Q9UBN0, Q9UCT0 | Gene names | SMR3B, PBII, PROL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog B precursor (Proline-rich protein 3) (Proline-rich peptide P-B) [Contains: Peptide P-A; Peptide D1A]. | |||||
|
DPP8_HUMAN
|
||||||
| NC score | 0.063738 (rank : 27) | θ value | 0.00869519 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6V1X1, Q7Z4C8, Q7Z4D3, Q7Z4E1, Q8IWG7, Q8NEM5, Q96JX1, Q9HBM2, Q9HBM3, Q9HBM4, Q9HBM5, Q9NXF4 | Gene names | DPP8, DPRP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 8 (EC 3.4.14.5) (Dipeptidyl peptidase VIII) (DP8) (Prolyl dipeptidase DPP8) (Dipeptidyl peptidase IV-related protein 1) (DPRP-1). | |||||
|
DPP8_MOUSE
|
||||||
| NC score | 0.061681 (rank : 28) | θ value | 0.00869519 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80YA7, Q9D4G6 | Gene names | Dpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 8 (EC 3.4.14.5) (Dipeptidyl peptidase VIII) (DP8). | |||||
|
DPP9_HUMAN
|
||||||
| NC score | 0.056368 (rank : 29) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86TI2, Q6AI37, Q6UAL0, Q6ZMT2, Q6ZNJ5, Q8N2J7, Q8N3F5, Q8WXD8, Q96NT8, Q9BVR3 | Gene names | DPP9, DPRP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9) (Dipeptidyl peptidase IV-related protein 2) (DPRP-2). | |||||
|
DPP9_MOUSE
|
||||||
| NC score | 0.049900 (rank : 30) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BVG4, Q6KAM9, Q8BWT9 | Gene names | Dpp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 9 (EC 3.4.14.5) (Dipeptidyl peptidase IX) (DP9) (Dipeptidyl peptidase-like protein 9) (DPLP9). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.044921 (rank : 31) | θ value | 0.00665767 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.044672 (rank : 32) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.038632 (rank : 33) | θ value | 0.365318 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.024800 (rank : 34) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.021521 (rank : 35) | θ value | 0.0431538 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.021282 (rank : 36) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
LAG3_MOUSE
|
||||||
| NC score | 0.015950 (rank : 37) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61790, Q0VBL2 | Gene names | Lag3 | |||
|
Domain Architecture |

|
|||||
| Description | Lymphocyte activation gene 3 protein precursor (LAG-3) (CD223 antigen). | |||||
|
BAG3_MOUSE
|
||||||
| NC score | 0.015897 (rank : 38) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JLV1, Q9JJC7 | Gene names | Bag3, Bis | |||
|
Domain Architecture |

|
|||||
| Description | BAG family molecular chaperone regulator 3 (BCL-2-binding athanogene- 3) (BAG-3) (Bcl-2-binding protein Bis). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.014888 (rank : 39) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.014794 (rank : 40) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.014613 (rank : 41) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.014195 (rank : 42) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
PLXB1_HUMAN
|
||||||
| NC score | 0.012421 (rank : 43) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43157, Q6NY20, Q9UIV7, Q9UJ92, Q9UJ93 | Gene names | PLXNB1, KIAA0407, SEP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor (Semaphorin receptor SEP). | |||||
|
GP108_MOUSE
|
||||||
| NC score | 0.011794 (rank : 44) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WD0, Q8BXE9, Q925B1 | Gene names | Gpr108, Lustr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein GPR108 precursor (Lung seven transmembrane receptor 2). | |||||
|
RKHD1_HUMAN
|
||||||
| NC score | 0.010374 (rank : 45) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86XN8 | Gene names | RKHD1, KIAA2031 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger and KH domain-containing protein 1. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.010268 (rank : 46) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
TRPV4_HUMAN
|
||||||
| NC score | 0.009702 (rank : 47) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HBA0, Q86YZ6, Q8NDY7, Q8NG64, Q96Q92, Q96RS7, Q9HBC0 | Gene names | TRPV4, VRL2, VROAC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily V member 4 (TrpV4) (osm-9-like TRP channel 4) (OTRPC4) (Vanilloid receptor-like channel 2) (Vanilloid receptor-like protein 2) (VRL-2) (Vanilloid receptor-related osmotically-activated channel) (VR-OAC) (Transient receptor potential protein 12) (TRP12). | |||||
|
EPN2_HUMAN
|
||||||
| NC score | 0.007682 (rank : 48) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95208, O95207, Q9H7Z2, Q9UPT7 | Gene names | EPN2, KIAA1065 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-2 (EPS-15-interacting protein 2). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.005897 (rank : 49) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.005438 (rank : 50) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.005327 (rank : 51) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
USH1C_MOUSE
|
||||||
| NC score | 0.003602 (rank : 52) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ES64, Q91XD1, Q9CVG7, Q9ES65 | Gene names | Ush1c | |||
|
Domain Architecture |

|
|||||
| Description | Harmonin (Usher syndrome type-1C protein homolog) (PDZ domain- containing protein). | |||||
|
ATP9B_MOUSE
|
||||||
| NC score | 0.003277 (rank : 53) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98195, Q99LI3 | Gene names | Atp9b | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase IIB (EC 3.6.3.1). | |||||
|
PTPRO_HUMAN
|
||||||
| NC score | 0.002950 (rank : 54) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16827, Q13101 | Gene names | PTPRO, GLEPP1, PTPU2 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase O precursor (EC 3.1.3.48) (Glomerular epithelial protein 1) (Protein tyrosine phosphatase U2) (PTPase U2) (PTP-U2). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.002429 (rank : 55) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | 0.002375 (rank : 56) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
TITIN_HUMAN
|
||||||
| NC score | -0.000315 (rank : 57) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | -0.000667 (rank : 58) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||