Please be patient as the page loads
|
EI2BD_HUMAN
|
||||||
| SwissProt Accessions | Q9UI10, Q5BJF4, Q9BUV9, Q9UBG4, Q9UIQ9, Q9UJ95 | Gene names | EIF2B4, EIF2BD | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EI2BD_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UI10, Q5BJF4, Q9BUV9, Q9UBG4, Q9UIQ9, Q9UJ95 | Gene names | EIF2B4, EIF2BD | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
EI2BD_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987486 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
EI2BA_MOUSE
|
||||||
| θ value | 5.79196e-23 (rank : 3) | NC score | 0.729860 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LC8 | Gene names | Eif2b1 | |||
|
Domain Architecture |
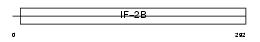
|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
EI2BA_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 4) | NC score | 0.725957 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14232 | Gene names | EIF2B1, EIF2BA | |||
|
Domain Architecture |
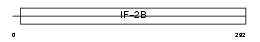
|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
EI2BB_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 5) | NC score | 0.652683 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49770, O43201 | Gene names | EIF2B2, EIF2BB | |||
|
Domain Architecture |
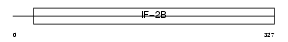
|
|||||
| Description | Translation initiation factor eIF-2B subunit beta (eIF-2B GDP-GTP exchange factor subunit beta) (S20I15) (S20III15). | |||||
|
EI2BB_MOUSE
|
||||||
| θ value | 6.20254e-17 (rank : 6) | NC score | 0.649929 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LD9 | Gene names | Eif2b2 | |||
|
Domain Architecture |
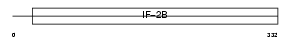
|
|||||
| Description | Translation initiation factor eIF-2B subunit beta (eIF-2B GDP-GTP exchange factor subunit beta). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.072096 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.044219 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZEP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.041599 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.024386 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ZN366_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.005343 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.051090 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.024020 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
CDC20_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.016818 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12834, Q9BW56, Q9UQI9 | Gene names | CDC20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC). | |||||
|
SPEG_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.011658 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.012097 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
UD14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.016115 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22310 | Gene names | UGT1A4, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-4 precursor (EC 2.4.1.17) (UDP- glucuronosyltransferase 1A4) (UDPGT) (UGT1*4) (UGT1-04) (UGT1.4) (UGT- 1D) (UGT1D) (Bilirubin-specific UDPGT isozyme 2) (HUG-BR2). | |||||
|
PPE2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.019062 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.018918 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.021929 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
LIMA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.021214 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
OXRP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.015261 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PHF3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.028143 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
UTS2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.045531 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZQ3 | Gene names | Uts2, Ucn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Urotensin-2 precursor (Urotensin-II) (U-II) (UII). | |||||
|
DDX49_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.005717 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.021099 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
M3K15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.002758 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 914 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZN16, Q5JPR4, Q6ZMV3 | Gene names | MAP3K15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 15 (EC 2.7.11.25) (MAPK/ERK kinase kinase 15) (MEK kinase 15) (MEKK 15). | |||||
|
HES3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.022345 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
KCNN3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.008894 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.005233 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.005520 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
UD13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.012476 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35503 | Gene names | UGT1A3, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-3 precursor (EC 2.4.1.17) (UDP- glucuronosyltransferase 1A3) (UDPGT) (UGT1*3) (UGT1-03) (UGT1.3) (UGT- 1C) (UGT1C). | |||||
|
MAVS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.017797 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
PLCE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.013013 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUQ2, Q8IZ47, Q9BQG4 | Gene names | AGPAT5 | |||
|
Domain Architecture |
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon (EC 2.3.1.51) (1-AGP acyltransferase 5) (1-AGPAT 5) (Lysophosphatidic acid acyltransferase-epsilon) (LPAAT-epsilon) (1-acylglycerol-3-phosphate O-acyltransferase 5). | |||||
|
EI2BD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UI10, Q5BJF4, Q9BUV9, Q9UBG4, Q9UIQ9, Q9UJ95 | Gene names | EIF2B4, EIF2BD | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
EI2BD_MOUSE
|
||||||
| NC score | 0.987486 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61749, Q61748, Q8VC35 | Gene names | Eif2b4, Eif2b, Eif2bd, Jgr1a | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit delta (eIF-2B GDP-GTP exchange factor subunit delta). | |||||
|
EI2BA_MOUSE
|
||||||
| NC score | 0.729860 (rank : 3) | θ value | 5.79196e-23 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LC8 | Gene names | Eif2b1 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
EI2BA_HUMAN
|
||||||
| NC score | 0.725957 (rank : 4) | θ value | 2.87452e-22 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14232 | Gene names | EIF2B1, EIF2BA | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit alpha (eIF-2B GDP-GTP exchange factor subunit alpha). | |||||
|
EI2BB_HUMAN
|
||||||
| NC score | 0.652683 (rank : 5) | θ value | 4.74913e-17 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P49770, O43201 | Gene names | EIF2B2, EIF2BB | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit beta (eIF-2B GDP-GTP exchange factor subunit beta) (S20I15) (S20III15). | |||||
|
EI2BB_MOUSE
|
||||||
| NC score | 0.649929 (rank : 6) | θ value | 6.20254e-17 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99LD9 | Gene names | Eif2b2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor eIF-2B subunit beta (eIF-2B GDP-GTP exchange factor subunit beta). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.072096 (rank : 7) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.051090 (rank : 8) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
UTS2_MOUSE
|
||||||
| NC score | 0.045531 (rank : 9) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9QZQ3 | Gene names | Uts2, Ucn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Urotensin-2 precursor (Urotensin-II) (U-II) (UII). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.044219 (rank : 10) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZEP1_MOUSE
|
||||||
| NC score | 0.041599 (rank : 11) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1063 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03172 | Gene names | Hivep1, Cryabp1, Znf40 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 40 (Transcription factor alphaA-CRYBP1) (Alpha A- crystallin-binding protein I) (Alpha A-CRYBP1). | |||||
|
PHF3_HUMAN
|
||||||
| NC score | 0.028143 (rank : 12) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92576, Q5T1T6, Q9NQ16, Q9UI45 | Gene names | PHF3, KIAA0244 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 3. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.024386 (rank : 13) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.024020 (rank : 14) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
HES3_MOUSE
|
||||||
| NC score | 0.022345 (rank : 15) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61657, O09083, O09150 | Gene names | Hes3, Hes-3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-3 (Hairy and enhancer of split 3). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.021929 (rank : 16) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
LIMA1_HUMAN
|
||||||
| NC score | 0.021214 (rank : 17) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.021099 (rank : 18) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
PPE2_MOUSE
|
||||||
| NC score | 0.019062 (rank : 19) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35385 | Gene names | Ppef2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase with EF-hands 2 (EC 3.1.3.16) (PPEF-2). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.018918 (rank : 20) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
MAVS_MOUSE
|
||||||
| NC score | 0.017797 (rank : 21) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VCF0 | Gene names | Mavs, Ips1, Visa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif). | |||||
|
CDC20_HUMAN
|
||||||
| NC score | 0.016818 (rank : 22) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12834, Q9BW56, Q9UQI9 | Gene names | CDC20 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 20 homolog (p55CDC). | |||||
|
UD14_HUMAN
|
||||||
| NC score | 0.016115 (rank : 23) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22310 | Gene names | UGT1A4, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-4 precursor (EC 2.4.1.17) (UDP- glucuronosyltransferase 1A4) (UDPGT) (UGT1*4) (UGT1-04) (UGT1.4) (UGT- 1D) (UGT1D) (Bilirubin-specific UDPGT isozyme 2) (HUG-BR2). | |||||
|
OXRP_HUMAN
|
||||||
| NC score | 0.015261 (rank : 24) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 544 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4L1 | Gene names | HYOU1, ORP150 | |||
|
Domain Architecture |

|
|||||
| Description | 150 kDa oxygen-regulated protein precursor (Orp150) (Hypoxia up- regulated 1). | |||||
|
PLCE_HUMAN
|
||||||
| NC score | 0.013013 (rank : 25) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NUQ2, Q8IZ47, Q9BQG4 | Gene names | AGPAT5 | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase epsilon (EC 2.3.1.51) (1-AGP acyltransferase 5) (1-AGPAT 5) (Lysophosphatidic acid acyltransferase-epsilon) (LPAAT-epsilon) (1-acylglycerol-3-phosphate O-acyltransferase 5). | |||||
|
UD13_HUMAN
|
||||||
| NC score | 0.012476 (rank : 26) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35503 | Gene names | UGT1A3, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-3 precursor (EC 2.4.1.17) (UDP- glucuronosyltransferase 1A3) (UDPGT) (UGT1*3) (UGT1-03) (UGT1.3) (UGT- 1C) (UGT1C). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.012097 (rank : 27) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
SPEG_HUMAN
|
||||||
| NC score | 0.011658 (rank : 28) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15772, Q27J74, Q695L1, Q6FGA6, Q6ZQW1, Q6ZTL8, Q9P2P9 | Gene names | SPEG, APEG1, KIAA1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle preferentially expressed protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
KCNN3_HUMAN
|
||||||
| NC score | 0.008894 (rank : 29) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGI6, O43517 | Gene names | KCNN3, K3 | |||
|
Domain Architecture |

|
|||||
| Description | Small conductance calcium-activated potassium channel protein 3 (SK3) (SKCa3). | |||||
|
DDX49_HUMAN
|
||||||
| NC score | 0.005717 (rank : 30) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6V7, Q53FJ1 | Gene names | DDX49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX49 (EC 3.6.1.-) (DEAD box protein 49). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.005520 (rank : 31) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
ZN366_HUMAN
|
||||||
| NC score | 0.005343 (rank : 32) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 732 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N895, Q7RTV4 | Gene names | ZNF366 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 366. | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.005233 (rank : 33) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
M3K15_HUMAN
|
||||||
| NC score | 0.002758 (rank : 34) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 914 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZN16, Q5JPR4, Q6ZMV3 | Gene names | MAP3K15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 15 (EC 2.7.11.25) (MAPK/ERK kinase kinase 15) (MEK kinase 15) (MEKK 15). | |||||