Please be patient as the page loads
|
DLGP2_MOUSE
|
||||||
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DLGP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979830 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14490, O14489, P78335 | Gene names | DLGAP1, DAP1, GKAP | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (hGKAP) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD- 95/SAP90-binding protein 1). | |||||
|
DLGP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.979571 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
DLGP2_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
DLGP3_HUMAN
|
||||||
| θ value | 7.01099e-170 (rank : 4) | NC score | 0.957506 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP3_MOUSE
|
||||||
| θ value | 1.52337e-140 (rank : 5) | NC score | 0.955757 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP4_HUMAN
|
||||||
| θ value | 1.74296e-136 (rank : 6) | NC score | 0.948310 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
DLG7_MOUSE
|
||||||
| θ value | 4.02038e-16 (rank : 7) | NC score | 0.596160 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
DLG7_HUMAN
|
||||||
| θ value | 5.8054e-15 (rank : 8) | NC score | 0.576765 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
CHK2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.003329 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O96017, Q6QA03, Q6QA04, Q6QA05, Q6QA06, Q6QA07, Q6QA08, Q6QA10, Q6QA11, Q6QA12, Q6QA13, Q9HCQ8, Q9UGF0, Q9UGF1 | Gene names | CHEK2, CHK2 | |||
|
Domain Architecture |
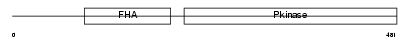
|
|||||
| Description | Serine/threonine-protein kinase Chk2 (EC 2.7.11.1) (Cds1). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.022622 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.023284 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CI079_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.031529 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
LRP12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.011362 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |
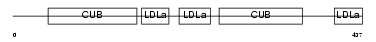
|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
PNPT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.032075 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
ANKS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.009324 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.003174 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
PTN22_MOUSE
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.008734 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.008812 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ERBB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.001769 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.027630 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.015022 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.002322 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.011040 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.005105 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
CCD55_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.009924 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.006845 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
XRN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.011639 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZH2, Q4G0S3, Q68D88, Q6AI24, Q6MZS8, Q86WS7, Q8N8U4, Q9UF39 | Gene names | XRN1, SEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (Strand-exchange protein 1 homolog). | |||||
|
ABLM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.003930 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.011365 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.007264 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
E2F3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.014428 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |
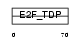
|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
E2F3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.011318 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
GPR75_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.011959 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95800, Q6NWR2 | Gene names | GPR75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 75. | |||||
|
GTSE1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.019692 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.008848 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.000690 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
PERL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.007242 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
|
STRAP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.003848 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1Z2 | Gene names | Strap, Unrip | |||
|
Domain Architecture |
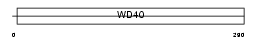
|
|||||
| Description | Serine-threonine kinase receptor-associated protein (UNR-interacting protein). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.002608 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.004344 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
HEM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.006312 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13196 | Gene names | ALAS1, ALASH | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
IRF3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.006300 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.010137 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.010144 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MYPT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.005005 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
PNPT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.022881 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
PO3F3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.004518 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20264, P78379 | Gene names | POU3F3, BRN1, OTF8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
PO3F3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.004512 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31361 | Gene names | Pou3f3, Brn-1, Brn1, Otf8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
TEX14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.004396 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
DLGP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
DLGP1_HUMAN
|
||||||
| NC score | 0.979830 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14490, O14489, P78335 | Gene names | DLGAP1, DAP1, GKAP | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (hGKAP) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD- 95/SAP90-binding protein 1). | |||||
|
DLGP1_MOUSE
|
||||||
| NC score | 0.979571 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D415, Q52KF6, Q5DTK5, Q6P6N4, Q6XBF4, Q8BZL7, Q8BZQ1, Q8C0G0 | Gene names | Dlgap1, Kiaa4162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 1 (DAP-1) (Guanylate kinase-associated protein) (SAP90/PSD-95-associated protein 1) (SAPAP1) (PSD-95/SAP90- binding protein 1). | |||||
|
DLGP3_HUMAN
|
||||||
| NC score | 0.957506 (rank : 4) | θ value | 7.01099e-170 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95886, Q5TDD5, Q9H3X7 | Gene names | DLGAP3, DAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP3_MOUSE
|
||||||
| NC score | 0.955757 (rank : 5) | θ value | 1.52337e-140 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6PFD5, Q6PDX0, Q6XBF2 | Gene names | Dlgap3, Dap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 3 (DAP-3) (SAP90/PSD-95-associated protein 3) (SAPAP3) (PSD-95/SAP90-binding protein 3). | |||||
|
DLGP4_HUMAN
|
||||||
| NC score | 0.948310 (rank : 6) | θ value | 1.74296e-136 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2H0, Q5T2Y4, Q5T2Y5, Q9H137, Q9H138, Q9H1L7 | Gene names | DLGAP4, DAP4, KIAA0964 | |||
|
Domain Architecture |

|
|||||
| Description | Disks large-associated protein 4 (DAP-4) (SAP90/PSD-95-associated protein 4) (SAPAP4) (PSD-95/SAP90-binding protein 4). | |||||
|
DLG7_MOUSE
|
||||||
| NC score | 0.596160 (rank : 7) | θ value | 4.02038e-16 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K4R9, Q6ZQL3, Q80VS7, Q8BUC8, Q8BZ79 | Gene names | Dlg7, Kiaa0008 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein homolog) (HURP). | |||||
|
DLG7_HUMAN
|
||||||
| NC score | 0.576765 (rank : 8) | θ value | 5.8054e-15 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15398, Q8NG58 | Gene names | DLG7, KIAA0008 | |||
|
Domain Architecture |

|
|||||
| Description | Discs large homolog 7 (Hepatoma up-regulated protein) (HURP). | |||||
|
PNPT1_HUMAN
|
||||||
| NC score | 0.032075 (rank : 9) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCS8, Q53SU0, Q68CN1, Q7Z7D1, Q8IWX1, Q96T05, Q9BRU3, Q9BVX0 | Gene names | PNPT1, PNPASE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.031529 (rank : 10) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.027630 (rank : 11) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.023284 (rank : 12) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
PNPT1_MOUSE
|
||||||
| NC score | 0.022881 (rank : 13) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1R3, Q3UEP9, Q810U7, Q812B3, Q8R2U3, Q9DC52 | Gene names | Pnpt1, Pnpase | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyribonucleotide nucleotidyltransferase 1, mitochondrial precursor (EC 2.7.7.8) (PNPase 1) (Polynucleotide phosphorylase-like protein) (PNPase old-35) (3'-5' RNA exonuclease OLD35). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.022622 (rank : 14) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
GTSE1_MOUSE
|
||||||
| NC score | 0.019692 (rank : 15) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R080, O89015, Q9CSG9 | Gene names | Gtse1, B99 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G2 and S phase expressed protein 1 (Gtse-1) (B99 protein). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.015022 (rank : 16) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
E2F3_HUMAN
|
||||||
| NC score | 0.014428 (rank : 17) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00716, Q15000, Q9BZ44 | Gene names | E2F3, KIAA0075 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
GPR75_HUMAN
|
||||||
| NC score | 0.011959 (rank : 18) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95800, Q6NWR2 | Gene names | GPR75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 75. | |||||
|
XRN1_HUMAN
|
||||||
| NC score | 0.011639 (rank : 19) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZH2, Q4G0S3, Q68D88, Q6AI24, Q6MZS8, Q86WS7, Q8N8U4, Q9UF39 | Gene names | XRN1, SEP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-3' exoribonuclease 1 (EC 3.1.11.-) (Strand-exchange protein 1 homolog). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.011365 (rank : 20) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
LRP12_HUMAN
|
||||||
| NC score | 0.011362 (rank : 21) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y561 | Gene names | LRP12, ST7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 12 precursor (Suppressor of tumorigenicity protein 7). | |||||
|
E2F3_MOUSE
|
||||||
| NC score | 0.011318 (rank : 22) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35261, Q5T0I6 | Gene names | E2f3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2F3 (E2F-3). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.011040 (rank : 23) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.010144 (rank : 24) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.010137 (rank : 25) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
CCD55_HUMAN
|
||||||
| NC score | 0.009924 (rank : 26) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0G5, Q6FI71 | Gene names | CCDC55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
ANKS1_HUMAN
|
||||||
| NC score | 0.009324 (rank : 27) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92625, Q5JYI9, Q5SYR2, Q86WQ7 | Gene names | ANKS1A, ANKS1, KIAA0229 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SAM domain-containing protein 1A (Odin). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.008848 (rank : 28) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.008812 (rank : 29) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
PTN22_MOUSE
|
||||||
| NC score | 0.008734 (rank : 30) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29352, Q7TMP9 | Gene names | Ptpn22, Ptpn8 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 22 (EC 3.1.3.48) (Hematopoietic cell protein-tyrosine phosphatase 70Z-PEP). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.007264 (rank : 31) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
PERL_HUMAN
|
||||||
| NC score | 0.007242 (rank : 32) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.006845 (rank : 33) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
HEM1_HUMAN
|
||||||
| NC score | 0.006312 (rank : 34) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13196 | Gene names | ALAS1, ALASH | |||
|
Domain Architecture |

|
|||||
| Description | 5-aminolevulinate synthase, nonspecific, mitochondrial precursor (EC 2.3.1.37) (5-aminolevulinic acid synthase) (Delta-aminolevulinate synthase) (Delta-ALA synthetase) (ALAS-H). | |||||
|
IRF3_HUMAN
|
||||||
| NC score | 0.006300 (rank : 35) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14653 | Gene names | IRF3 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon regulatory factor 3 (IRF-3). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.005105 (rank : 36) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
MYPT2_HUMAN
|
||||||
| NC score | 0.005005 (rank : 37) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 707 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60237, Q8N179, Q9HCB7, Q9HCB8 | Gene names | PPP1R12B, MYPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12B (Myosin phosphatase- targeting subunit 2) (Myosin phosphatase target subunit 2). | |||||
|
PO3F3_HUMAN
|
||||||
| NC score | 0.004518 (rank : 38) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20264, P78379 | Gene names | POU3F3, BRN1, OTF8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
PO3F3_MOUSE
|
||||||
| NC score | 0.004512 (rank : 39) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P31361 | Gene names | Pou3f3, Brn-1, Brn1, Otf8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
TEX14_HUMAN
|
||||||
| NC score | 0.004396 (rank : 40) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1103 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IWB6, Q7RTP3, Q8ND97, Q9BXT9 | Gene names | TEX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.004344 (rank : 41) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
ABLM3_HUMAN
|
||||||
| NC score | 0.003930 (rank : 42) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
STRAP_MOUSE
|
||||||
| NC score | 0.003848 (rank : 43) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z1Z2 | Gene names | Strap, Unrip | |||
|
Domain Architecture |

|
|||||
| Description | Serine-threonine kinase receptor-associated protein (UNR-interacting protein). | |||||
|
CHK2_HUMAN
|
||||||
| NC score | 0.003329 (rank : 44) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O96017, Q6QA03, Q6QA04, Q6QA05, Q6QA06, Q6QA07, Q6QA08, Q6QA10, Q6QA11, Q6QA12, Q6QA13, Q9HCQ8, Q9UGF0, Q9UGF1 | Gene names | CHEK2, CHK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Chk2 (EC 2.7.11.1) (Cds1). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.003174 (rank : 45) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.002608 (rank : 46) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.002322 (rank : 47) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ERBB3_HUMAN
|
||||||
| NC score | 0.001769 (rank : 48) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.000690 (rank : 49) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||