Please be patient as the page loads
|
CD44_MOUSE
|
||||||
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CD44_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.972148 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |
|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
CD44_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
|
XLKD1_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 3) | NC score | 0.608778 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
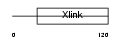
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 4) | NC score | 0.584021 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
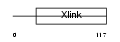
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
TSG6_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 5) | NC score | 0.307729 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 6) | NC score | 0.305405 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 7) | NC score | 0.180972 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 8) | NC score | 0.173974 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 9) | NC score | 0.181827 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 10) | NC score | 0.188112 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PGCB_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 11) | NC score | 0.199857 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
HPLN1_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.239033 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
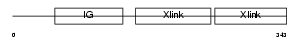
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.239374 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 19 | |
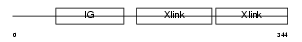
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |
|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.209284 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
CSPG2_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.173331 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
HPLN3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.237689 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |
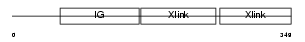
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
CN037_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.083084 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.044342 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.063984 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.061362 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
MUC15_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.077276 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
CSPG3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.156144 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
HPLN3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.238973 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |
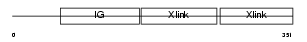
|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.054891 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.034349 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
PGCB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.182770 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.154242 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.044638 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NCA11_MOUSE
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.015723 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
NUFP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.042498 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5F2E7, Q3TCE2, Q3V195, Q80TF1 | Gene names | Nufip2, Kiaa1321 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.042810 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BCL9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.046391 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
HIPK2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.006395 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZR5, O88905, Q99P45, Q99P46, Q9D2E6, Q9D474, Q9EQL2, Q9QZR4 | Gene names | Hipk2, Nbak1, Stank | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (Nuclear body- associated kinase 1) (Sialophorin tail-associated nuclear serine/threonine-protein kinase). | |||||
|
KI13A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.016922 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
MYCD_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.030163 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
RSNL2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.027500 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CI96, Q8BW09, Q921Q4, Q9D2S6 | Gene names | Rsnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin-like protein 2. | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.051483 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.052038 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
KC1G1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 39) | NC score | 0.009067 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTH8 | Gene names | Csnk1g1 | |||
|
Domain Architecture |
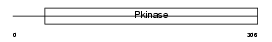
|
|||||
| Description | Casein kinase I isoform gamma-1 (EC 2.7.11.1) (CKI-gamma 1). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.059527 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
KI13A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.015597 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
NU214_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.047809 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PTN12_MOUSE
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.011867 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
TACT_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.044315 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40200 | Gene names | CD96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell surface protein tactile precursor (T cell-activated increased late expression protein) (CD96 antigen). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.042368 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
PTN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.008065 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.009538 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
HXA11_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.005981 (rank : 72) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
LR37A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.031757 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
LR37B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.022874 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.015216 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
ELOA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.014116 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
HN1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.023158 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PGH2 | Gene names | D17Ertd441e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1-like protein (HN1-like protein). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.011622 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
LOXE3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.007764 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYJ1, Q9H4F2, Q9HC22 | Gene names | ALOXE3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
METK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.017971 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00266, Q5QP09 | Gene names | MAT1A, AMS1, MATA1 | |||
|
Domain Architecture |
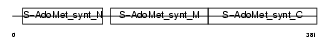
|
|||||
| Description | S-adenosylmethionine synthetase isoform type-1 (EC 2.5.1.6) (Methionine adenosyltransferase 1) (AdoMet synthetase 1) (Methionine adenosyltransferase I/III) (MAT-I/III). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.019692 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.003258 (rank : 75) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ADA22_MOUSE
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.004791 (rank : 73) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1V6, Q5TLI8, Q5TLI9, Q5TLJ0, Q5TLJ1, Q5TLJ2, Q5TLJ3, Q5TLJ4, Q5TLJ5, Q5TLJ6, Q5TLJ7, Q5TLJ8, Q5TLJ9, Q5TLK0, Q5TLK1, Q5TLK2, Q5TLK3, Q5TLK4, Q5TLK5, Q5TLK6, Q5TLK7, Q5TLK8, Q8BSF2, Q9R1V5 | Gene names | Adam22 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 22 precursor (A disintegrin and metalloproteinase domain 22). | |||||
|
CLAP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.016001 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CLAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.016050 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
DEAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.011989 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
METK1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.016625 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X83 | Gene names | Mat1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-adenosylmethionine synthetase isoform type-1 (EC 2.5.1.6) (Methionine adenosyltransferase 1) (AdoMet synthetase 1). | |||||
|
NR1D2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.003905 (rank : 74) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60674, Q60646 | Gene names | Nr1d2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (Orphan nuclear receptor RVR). | |||||
|
CLC4D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.052034 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
HPLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.212187 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |
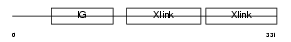
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.211123 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |
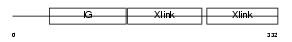
|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.209282 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |
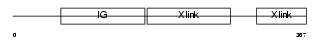
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.206611 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |
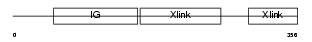
|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
LCE1D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.064466 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
LORI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.083404 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
REG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051145 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
REG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055654 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.113921 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.113644 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
CD44_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P15379, Q05732, Q61395, Q62060, Q62061, Q62062, Q62063, Q62408, Q62409, Q64296, Q99J14, Q9QYX8 | Gene names | Cd44, Ly-24 | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein 1) (PGP-1) (HUTCH-I) (Extracellular matrix receptor III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Lymphocyte antigen Ly-24). | |||||
|
CD44_HUMAN
|
||||||
| NC score | 0.972148 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16070, P22511, Q04858, Q13419, Q13957, Q13958, Q13959, Q13960, Q13961, Q13967, Q13968, Q13980, Q15861, Q16064, Q16065, Q16066, Q16208, Q16522, Q96J24 | Gene names | CD44, LHR | |||
|
Domain Architecture |

|
|||||
| Description | CD44 antigen precursor (Phagocytic glycoprotein I) (PGP-1) (HUTCH-I) (Extracellular matrix receptor-III) (ECMR-III) (GP90 lymphocyte homing/adhesion receptor) (Hermes antigen) (Hyaluronate receptor) (Heparan sulfate proteoglycan) (Epican) (CDw44). | |||||
|
XLKD1_HUMAN
|
||||||
| NC score | 0.608778 (rank : 3) | θ value | 8.95645e-16 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y5Y7, Q8TC18, Q9UNF4 | Gene names | XLKD1, CRSBP1, HAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Hyaluronic acid receptor) (Extracellular link domain-containing protein 1). | |||||
|
XLKD1_MOUSE
|
||||||
| NC score | 0.584021 (rank : 4) | θ value | 2.69671e-12 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BHC0, Q3TUC1, Q99NE4 | Gene names | Xlkd1, Crsbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphatic vessel endothelial hyaluronic acid receptor 1 precursor (LYVE-1) (Cell surface retention sequence-binding protein 1) (CRSBP-1) (Extracellular link domain-containing protein 1). | |||||
|
TSG6_HUMAN
|
||||||
| NC score | 0.307729 (rank : 5) | θ value | 1.06045e-08 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P98066, Q8WWI9 | Gene names | TNFAIP6, TSG6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein) (Hyaluronate-binding protein). | |||||
|
TSG6_MOUSE
|
||||||
| NC score | 0.305405 (rank : 6) | θ value | 1.80886e-08 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08859 | Gene names | Tnfaip6, Tnfip6, Tsg6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor-inducible protein TSG-6 precursor (TNF- stimulated gene 6 protein). | |||||
|
HPLN1_MOUSE
|
||||||
| NC score | 0.239374 (rank : 7) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QUP5, Q9D1G9, Q9Z1X7 | Gene names | Hapln1, Crtl1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN1_HUMAN
|
||||||
| NC score | 0.239033 (rank : 8) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10915 | Gene names | HAPLN1, CRTL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 1 precursor (Proteoglycan link protein) (Cartilage link protein) (LP). | |||||
|
HPLN3_HUMAN
|
||||||
| NC score | 0.238973 (rank : 9) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96S86 | Gene names | HAPLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor. | |||||
|
HPLN3_MOUSE
|
||||||
| NC score | 0.237689 (rank : 10) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q80WM5, Q80XX3 | Gene names | Hapln3, Lpr3 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 3 precursor (Link protein 3). | |||||
|
HPLN2_HUMAN
|
||||||
| NC score | 0.212187 (rank : 11) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9GZV7 | Gene names | HAPLN2, BRAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
HPLN2_MOUSE
|
||||||
| NC score | 0.211123 (rank : 12) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9ESM3 | Gene names | Hapln2, Bral1 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 2 precursor (Brain link protein 1). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.209284 (rank : 13) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
HPLN4_HUMAN
|
||||||
| NC score | 0.209282 (rank : 14) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86UW8, Q96PW2 | Gene names | HAPLN4, BRAL2, KIAA1926 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2). | |||||
|
HPLN4_MOUSE
|
||||||
| NC score | 0.206611 (rank : 15) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q80WM4, Q80XX2 | Gene names | Hapln4, Bral2, Lpr4 | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan and proteoglycan link protein 4 precursor (Brain link protein 2) (Link protein 4). | |||||
|
PGCB_MOUSE
|
||||||
| NC score | 0.199857 (rank : 16) | θ value | 0.00175202 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q61361 | Gene names | Bcan | |||
|
Domain Architecture |

|
|||||
| Description | Brevican core protein precursor. | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.188112 (rank : 17) | θ value | 0.00134147 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PGCB_HUMAN
|
||||||
| NC score | 0.182770 (rank : 18) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96GW7, Q8TBB9, Q9HBK1, Q9HBK4 | Gene names | BCAN, BEHAB, CSPG7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brevican core protein precursor (Brain-enriched hyaluronan-binding protein) (Protein BEHAB). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.181827 (rank : 19) | θ value | 0.00035302 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.180972 (rank : 20) | θ value | 5.26297e-08 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.173974 (rank : 21) | θ value | 1.69304e-06 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
CSPG2_HUMAN
|
||||||
| NC score | 0.173331 (rank : 22) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13611, P20754, Q13010, Q13189, Q15123, Q9UCL9, Q9UNW5 | Gene names | CSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M) (Glial hyaluronate-binding protein) (GHAP). | |||||
|
CSPG3_HUMAN
|
||||||
| NC score | 0.156144 (rank : 23) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14594, Q9UPK6 | Gene names | CSPG3, NCAN, NEUR | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.154242 (rank : 24) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.113921 (rank : 25) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.113644 (rank : 26) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
LORI_MOUSE
|
||||||
| NC score | 0.083404 (rank : 27) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P18165, Q8R0I9 | Gene names | Lor | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Loricrin. | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.083084 (rank : 28) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
MUC15_HUMAN
|
||||||
| NC score | 0.077276 (rank : 29) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N387, Q6UWS3, Q8IXI8, Q8WW41 | Gene names | MUC15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-15 precursor. | |||||
|
LCE1D_HUMAN
|
||||||
| NC score | 0.064466 (rank : 30) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5T752 | Gene names | LCE1D, LEP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 1D (Late envelope protein 4). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.063984 (rank : 31) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.061362 (rank : 32) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.059527 (rank : 33) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
REG4_MOUSE
|
||||||
| NC score | 0.055654 (rank : 34) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8G5, Q3TS25, Q9D858 | Gene names | Reg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.054891 (rank : 35) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.052038 (rank : 36) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
CLC4D_HUMAN
|
||||||
| NC score | 0.052034 (rank : 37) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WXI8, Q8N5J5 | Gene names | CLEC4D, CLECSF8, MCL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 4 member D (C-type lectin superfamily member 8) (C-type lectin-like receptor 6) (CLEC-6). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.051483 (rank : 38) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
REG4_HUMAN
|
||||||
| NC score | 0.051145 (rank : 39) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BYZ8, Q8NER6, Q8NER7 | Gene names | REG4, GISP, RELP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regenerating islet-derived protein 4 precursor (Reg IV) (REG-like protein) (Gastrointestinal secretory protein). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.047809 (rank : 40) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
BCL9_MOUSE
|
||||||
| NC score | 0.046391 (rank : 41) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D219, Q67FX9, Q8BUJ8, Q8VE74 | Gene names | Bcl9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.044638 (rank : 42) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.044342 (rank : 43) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
TACT_HUMAN
|
||||||
| NC score | 0.044315 (rank : 44) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P40200 | Gene names | CD96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell surface protein tactile precursor (T cell-activated increased late expression protein) (CD96 antigen). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.042810 (rank : 45) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
NUFP2_MOUSE
|
||||||
| NC score | 0.042498 (rank : 46) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5F2E7, Q3TCE2, Q3V195, Q80TF1 | Gene names | Nufip2, Kiaa1321 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear fragile X mental retardation-interacting protein 2 (FMRP- interacting protein 2) (82 kDa FMRP-interacting protein) (82-FIP). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.042368 (rank : 47) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.034349 (rank : 48) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
LR37A_HUMAN
|
||||||
| NC score | 0.031757 (rank : 49) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60309, Q49A01, Q49A80, Q8NB33 | Gene names | LRRC37A, KIAA0563 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37A. | |||||
|
MYCD_MOUSE
|
||||||
| NC score | 0.030163 (rank : 50) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VIM5, Q8C3W6, Q8VIL4 | Gene names | Myocd, Bsac2, Mycd, Srfcp | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin (SRF cofactor protein) (Basic SAP coiled-coil transcription activator 2). | |||||
|
RSNL2_MOUSE
|
||||||
| NC score | 0.027500 (rank : 51) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CI96, Q8BW09, Q921Q4, Q9D2S6 | Gene names | Rsnl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin-like protein 2. | |||||
|
HN1L_MOUSE
|
||||||
| NC score | 0.023158 (rank : 52) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PGH2 | Gene names | D17Ertd441e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1-like protein (HN1-like protein). | |||||
|
LR37B_HUMAN
|
||||||
| NC score | 0.022874 (rank : 53) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96QE4, Q5YKG6 | Gene names | LRRC37B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 37B precursor (C66 SLIT-like testicular protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.019692 (rank : 54) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
METK1_HUMAN
|
||||||
| NC score | 0.017971 (rank : 55) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00266, Q5QP09 | Gene names | MAT1A, AMS1, MATA1 | |||
|
Domain Architecture |
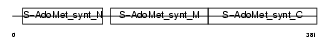
|
|||||
| Description | S-adenosylmethionine synthetase isoform type-1 (EC 2.5.1.6) (Methionine adenosyltransferase 1) (AdoMet synthetase 1) (Methionine adenosyltransferase I/III) (MAT-I/III). | |||||
|
KI13A_HUMAN
|
||||||
| NC score | 0.016922 (rank : 56) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1H9, Q9H1H8 | Gene names | KIF13A, RBKIN | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A (Kinesin-like protein RBKIN). | |||||
|
METK1_MOUSE
|
||||||
| NC score | 0.016625 (rank : 57) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X83 | Gene names | Mat1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-adenosylmethionine synthetase isoform type-1 (EC 2.5.1.6) (Methionine adenosyltransferase 1) (AdoMet synthetase 1). | |||||
|
CLAP2_MOUSE
|
||||||
| NC score | 0.016050 (rank : 58) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BRT1, Q8CHE3, Q99JI3, Q9DB80 | Gene names | Clasp2, Kiaa0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
CLAP2_HUMAN
|
||||||
| NC score | 0.016001 (rank : 59) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75122, Q7L8F6, Q8N6R6, Q9BQT3, Q9BQT4, Q9H7A3, Q9NSZ2 | Gene names | CLASP2, KIAA0627 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CLIP-associating protein 2 (Cytoplasmic linker-associated protein 2). | |||||
|
NCA11_MOUSE
|
||||||
| NC score | 0.015723 (rank : 60) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P13595, Q61949 | Gene names | Ncam1, Ncam | |||
|
Domain Architecture |

|
|||||
| Description | Neural cell adhesion molecule 1, 180 kDa isoform precursor (N-CAM 180) (NCAM-180). | |||||
|
KI13A_MOUSE
|
||||||
| NC score | 0.015597 (rank : 61) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EQW7, O35062 | Gene names | Kif13a | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF13A. | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.015216 (rank : 62) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
ELOA1_HUMAN
|
||||||
| NC score | 0.014116 (rank : 63) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14241, Q8IXH1 | Gene names | TCEB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor B polypeptide 3 (RNA polymerase II transcription factor SIII subunit A1) (SIII p110) (Elongin-A) (EloA) (Elongin 110 kDa subunit). | |||||
|
DEAF1_HUMAN
|
||||||
| NC score | 0.011989 (rank : 64) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75398, O15152, O75399, O75510, O75511, O75512, O75513, Q9UET1 | Gene names | DEAF1, SPN, ZMYND5 | |||
|
Domain Architecture |

|
|||||
| Description | Deformed epidermal autoregulatory factor 1 homolog (Nuclear DEAF-1- related transcriptional regulator) (NUDR) (Suppressin) (Zinc finger MYND domain-containing protein 5). | |||||
|
PTN12_MOUSE
|
||||||
| NC score | 0.011867 (rank : 65) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35831 | Gene names | Ptpn12 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 12 (EC 3.1.3.48) (Protein-tyrosine phosphatase P19) (P19-PTP) (MPTP-PEST). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.011622 (rank : 66) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.009538 (rank : 67) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
KC1G1_MOUSE
|
||||||
| NC score | 0.009067 (rank : 68) | θ value | 1.38821 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BTH8 | Gene names | Csnk1g1 | |||
|
Domain Architecture |

|
|||||
| Description | Casein kinase I isoform gamma-1 (EC 2.7.11.1) (CKI-gamma 1). | |||||
|
PTN4_HUMAN
|
||||||
| NC score | 0.008065 (rank : 69) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P29074 | Gene names | PTPN4 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 4 (EC 3.1.3.48) (Protein-tyrosine phosphatase MEG1) (PTPase-MEG1) (MEG). | |||||
|
LOXE3_HUMAN
|
||||||
| NC score | 0.007764 (rank : 70) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYJ1, Q9H4F2, Q9HC22 | Gene names | ALOXE3 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermis-type lipoxygenase 3 (EC 1.13.11.-) (e-LOX-3). | |||||
|
HIPK2_MOUSE
|
||||||
| NC score | 0.006395 (rank : 71) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZR5, O88905, Q99P45, Q99P46, Q9D2E6, Q9D474, Q9EQL2, Q9QZR4 | Gene names | Hipk2, Nbak1, Stank | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 2 (EC 2.7.11.1) (Nuclear body- associated kinase 1) (Sialophorin tail-associated nuclear serine/threonine-protein kinase). | |||||
|
HXA11_HUMAN
|
||||||
| NC score | 0.005981 (rank : 72) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31270 | Gene names | HOXA11, HOX1I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A11 (Hox-1I). | |||||
|
ADA22_MOUSE
|
||||||
| NC score | 0.004791 (rank : 73) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R1V6, Q5TLI8, Q5TLI9, Q5TLJ0, Q5TLJ1, Q5TLJ2, Q5TLJ3, Q5TLJ4, Q5TLJ5, Q5TLJ6, Q5TLJ7, Q5TLJ8, Q5TLJ9, Q5TLK0, Q5TLK1, Q5TLK2, Q5TLK3, Q5TLK4, Q5TLK5, Q5TLK6, Q5TLK7, Q5TLK8, Q8BSF2, Q9R1V5 | Gene names | Adam22 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 22 precursor (A disintegrin and metalloproteinase domain 22). | |||||
|
NR1D2_MOUSE
|
||||||
| NC score | 0.003905 (rank : 74) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60674, Q60646 | Gene names | Nr1d2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (Orphan nuclear receptor RVR). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.003258 (rank : 75) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||