Please be patient as the page loads
|
SPY1_HUMAN
|
||||||
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPY1_HUMAN
|
||||||
| θ value | 4.39142e-172 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY1_MOUSE
|
||||||
| θ value | 8.93083e-141 (rank : 2) | NC score | 0.987127 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QXV9 | Gene names | Spry1 | |||
|
Domain Architecture |
|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY2_HUMAN
|
||||||
| θ value | 2.47765e-74 (rank : 3) | NC score | 0.954425 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| θ value | 2.3998e-69 (rank : 4) | NC score | 0.952514 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY4_HUMAN
|
||||||
| θ value | 1.66964e-54 (rank : 5) | NC score | 0.937281 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9C004, Q6QIX2, Q9C003 | Gene names | SPRY4 | |||
|
Domain Architecture |
|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY4_MOUSE
|
||||||
| θ value | 6.34462e-54 (rank : 6) | NC score | 0.935870 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |
|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY3_HUMAN
|
||||||
| θ value | 2.49495e-50 (rank : 7) | NC score | 0.943757 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43610 | Gene names | SPRY3 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 3 (Spry-3). | |||||
|
SPRE1_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 8) | NC score | 0.509407 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE1_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 9) | NC score | 0.508388 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q924S8, Q6PET8 | Gene names | Spred1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE2_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 10) | NC score | 0.507863 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE3_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 11) | NC score | 0.516723 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE3_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 12) | NC score | 0.508247 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE2_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 13) | NC score | 0.503587 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
DLL3_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 14) | NC score | 0.064557 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
ITB3_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 15) | NC score | 0.085727 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
ITB3_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 16) | NC score | 0.084411 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.083234 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
ITB2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.070071 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.051686 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
TSP4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.052923 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35443, Q86TG2 | Gene names | THBS4, TSP4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
USH2A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.046521 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.060494 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.063056 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.079563 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
TSP4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.046502 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z1T2, Q9QYS3, Q9WUE0 | Gene names | Thbs4, Tsp4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.045202 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
ATRN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.047871 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
NOTC2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 28) | NC score | 0.043908 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
KR106_HUMAN
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.058072 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.074603 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.045864 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
IGF1R_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.004783 (rank : 86) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
KR108_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.060471 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.013597 (rank : 81) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.052292 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.055344 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.018796 (rank : 79) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.042751 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.046267 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZFY21_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.021164 (rank : 78) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.039763 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.048555 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ITB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.081305 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.053628 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
SFR14_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.012405 (rank : 82) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
WDFY2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.011447 (rank : 83) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BUB4 | Gene names | Wdfy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2). | |||||
|
ZFY21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.022973 (rank : 76) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.045232 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
KR511_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.071989 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.052273 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.034302 (rank : 73) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.043280 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.032105 (rank : 75) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
VEGFC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.021947 (rank : 77) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49767 | Gene names | VEGFC | |||
|
Domain Architecture |
|
|||||
| Description | Vascular endothelial growth factor C precursor (VEGF-C) (Vascular endothelial growth factor-related protein) (VRP) (Flt4 ligand) (Flt4- L). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.041014 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
GPC6A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.003996 (rank : 87) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.006201 (rank : 85) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.046635 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR410_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.032773 (rank : 74) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.051450 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.069068 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.036462 (rank : 72) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LRP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.016926 (rank : 80) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
WDFY2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010103 (rank : 84) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P53, Q96CS1 | Gene names | WDFY2, WDF2, ZFYVE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2) (Zinc finger FYVE domain-containing protein 22). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.066384 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.069218 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EVL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.085662 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
EVL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.081890 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ITB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.056592 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.051628 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.053460 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
ITB5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.053004 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
ITB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.055414 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.054972 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.054269 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P26010 | Gene names | ITGB7 | |||
|
Domain Architecture |
|
|||||
| Description | Integrin beta-7 precursor. | |||||
|
ITB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053299 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P26011, Q3U2M1, Q64656 | Gene names | Itgb7 | |||
|
Domain Architecture |
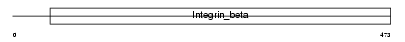
|
|||||
| Description | Integrin beta-7 precursor (Integrin beta-P) (M290 IEL antigen). | |||||
|
ITB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.059848 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
KR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.095042 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.052534 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.079817 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.062720 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.058198 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.056763 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
MT4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.050684 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47945 | Gene names | Mt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-4 (MT-4) (Metallothionein-IV) (MT-IV). | |||||
|
SREC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.054259 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
VASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.079871 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.083811 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
SPY1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.39142e-172 (rank : 1) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY1_MOUSE
|
||||||
| NC score | 0.987127 (rank : 2) | θ value | 8.93083e-141 (rank : 2) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9QXV9 | Gene names | Spry1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY2_HUMAN
|
||||||
| NC score | 0.954425 (rank : 3) | θ value | 2.47765e-74 (rank : 3) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| NC score | 0.952514 (rank : 4) | θ value | 2.3998e-69 (rank : 4) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY3_HUMAN
|
||||||
| NC score | 0.943757 (rank : 5) | θ value | 2.49495e-50 (rank : 7) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O43610 | Gene names | SPRY3 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 3 (Spry-3). | |||||
|
SPY4_HUMAN
|
||||||
| NC score | 0.937281 (rank : 6) | θ value | 1.66964e-54 (rank : 5) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9C004, Q6QIX2, Q9C003 | Gene names | SPRY4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY4_MOUSE
|
||||||
| NC score | 0.935870 (rank : 7) | θ value | 6.34462e-54 (rank : 6) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPRE3_HUMAN
|
||||||
| NC score | 0.516723 (rank : 8) | θ value | 8.11959e-09 (rank : 11) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE1_HUMAN
|
||||||
| NC score | 0.509407 (rank : 9) | θ value | 1.25267e-09 (rank : 8) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE1_MOUSE
|
||||||
| NC score | 0.508388 (rank : 10) | θ value | 1.25267e-09 (rank : 9) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q924S8, Q6PET8 | Gene names | Spred1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE3_MOUSE
|
||||||
| NC score | 0.508247 (rank : 11) | θ value | 8.11959e-09 (rank : 12) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE2_HUMAN
|
||||||
| NC score | 0.507863 (rank : 12) | θ value | 8.11959e-09 (rank : 10) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE2_MOUSE
|
||||||
| NC score | 0.503587 (rank : 13) | θ value | 1.06045e-08 (rank : 13) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.095042 (rank : 14) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
ITB3_MOUSE
|
||||||
| NC score | 0.085727 (rank : 15) | θ value | 0.0193708 (rank : 15) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.085662 (rank : 16) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ITB3_HUMAN
|
||||||
| NC score | 0.084411 (rank : 17) | θ value | 0.0252991 (rank : 16) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.083811 (rank : 18) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.083234 (rank : 19) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.081890 (rank : 20) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.081305 (rank : 21) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.079871 (rank : 22) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.079817 (rank : 23) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.079563 (rank : 24) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.074603 (rank : 25) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.071989 (rank : 26) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
ITB2_HUMAN
|
||||||
| NC score | 0.070071 (rank : 27) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.069218 (rank : 28) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.069068 (rank : 29) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.066384 (rank : 30) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
DLL3_HUMAN
|
||||||
| NC score | 0.064557 (rank : 31) | θ value | 0.0193708 (rank : 14) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NYJ7 | Gene names | DLL3 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 3 precursor (Drosophila Delta homolog 3). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.063056 (rank : 32) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.062720 (rank : 33) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.060494 (rank : 34) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.060471 (rank : 35) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
ITB8_HUMAN
|
||||||
| NC score | 0.059848 (rank : 36) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.058198 (rank : 37) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.058072 (rank : 38) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.056763 (rank : 39) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
ITB1_HUMAN
|
||||||
| NC score | 0.056592 (rank : 40) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P05556, P78466, P78467, Q13089, Q13090, Q13091, Q13212, Q14622, Q14647 | Gene names | ITGB1, FNRB | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
ITB6_HUMAN
|
||||||
| NC score | 0.055414 (rank : 41) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P18564, Q16500 | Gene names | ITGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.055344 (rank : 42) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
ITB6_MOUSE
|
||||||
| NC score | 0.054972 (rank : 43) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Z0T9 | Gene names | Itgb6 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-6 precursor. | |||||
|
ITB7_HUMAN
|
||||||
| NC score | 0.054269 (rank : 44) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P26010 | Gene names | ITGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-7 precursor. | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.054259 (rank : 45) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.053628 (rank : 46) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
ITB5_HUMAN
|
||||||
| NC score | 0.053460 (rank : 47) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P18084 | Gene names | ITGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
ITB7_MOUSE
|
||||||
| NC score | 0.053299 (rank : 48) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P26011, Q3U2M1, Q64656 | Gene names | Itgb7 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-7 precursor (Integrin beta-P) (M290 IEL antigen). | |||||
|
ITB5_MOUSE
|
||||||
| NC score | 0.053004 (rank : 49) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
TSP4_HUMAN
|
||||||
| NC score | 0.052923 (rank : 50) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P35443, Q86TG2 | Gene names | THBS4, TSP4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.052534 (rank : 51) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.052292 (rank : 52) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.052273 (rank : 53) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.051686 (rank : 54) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ITB1_MOUSE
|
||||||
| NC score | 0.051628 (rank : 55) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P09055 | Gene names | Itgb1 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-1 precursor (Fibronectin receptor subunit beta) (Integrin VLA-4 subunit beta) (CD29 antigen). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.051450 (rank : 56) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
MT4_MOUSE
|
||||||
| NC score | 0.050684 (rank : 57) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P47945 | Gene names | Mt4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-4 (MT-4) (Metallothionein-IV) (MT-IV). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.048555 (rank : 58) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.047871 (rank : 59) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.046635 (rank : 60) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.046521 (rank : 61) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
TSP4_MOUSE
|
||||||
| NC score | 0.046502 (rank : 62) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z1T2, Q9QYS3, Q9WUE0 | Gene names | Thbs4, Tsp4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.046267 (rank : 63) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.045864 (rank : 64) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.045232 (rank : 65) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.045202 (rank : 66) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
NOTC2_MOUSE
|
||||||
| NC score | 0.043908 (rank : 67) | θ value | 0.813845 (rank : 28) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1069 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35516, Q06008, Q60941 | Gene names | Notch2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (Motch B) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.043280 (rank : 68) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.042751 (rank : 69) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.041014 (rank : 70) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.039763 (rank : 71) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.036462 (rank : 72) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.034302 (rank : 73) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
KR410_HUMAN
|
||||||
| NC score | 0.032773 (rank : 74) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ7 | Gene names | KRTAP4-10, KAP4.10, KRTAP4.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 4-10 (Keratin-associated protein 4.10) (Ultrahigh sulfur keratin-associated protein 4.10). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.032105 (rank : 75) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
ZFY21_MOUSE
|
||||||
| NC score | 0.022973 (rank : 76) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VCM3, Q9D1E2 | Gene names | Zfyve21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
VEGFC_HUMAN
|
||||||
| NC score | 0.021947 (rank : 77) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49767 | Gene names | VEGFC | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor C precursor (VEGF-C) (Vascular endothelial growth factor-related protein) (VRP) (Flt4 ligand) (Flt4- L). | |||||
|
ZFY21_HUMAN
|
||||||
| NC score | 0.021164 (rank : 78) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQ24, Q86T05, Q96LT1 | Gene names | ZFYVE21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger FYVE domain-containing protein 21. | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.018796 (rank : 79) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
LRP4_MOUSE
|
||||||
| NC score | 0.016926 (rank : 80) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8VI56, Q8BPX5, Q8CBB3, Q8CCP5 | Gene names | Lrp4, Kiaa0816 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 4 precursor (LDLR dan). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.013597 (rank : 81) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
SFR14_MOUSE
|
||||||
| NC score | 0.012405 (rank : 82) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8CH09, Q6PG19, Q80UY8, Q8BY32, Q8CFM0 | Gene names | Sfrs14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative splicing factor, arginine/serine-rich 14 (Arginine/serine- rich-splicing factor 14). | |||||
|
WDFY2_MOUSE
|
||||||
| NC score | 0.011447 (rank : 83) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BUB4 | Gene names | Wdfy2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2). | |||||
|
WDFY2_HUMAN
|
||||||
| NC score | 0.010103 (rank : 84) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96P53, Q96CS1 | Gene names | WDFY2, WDF2, ZFYVE22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 2 (WD40- and FYVE domain- containing protein 2) (Zinc finger FYVE domain-containing protein 22). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.006201 (rank : 85) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
IGF1R_HUMAN
|
||||||
| NC score | 0.004783 (rank : 86) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
GPC6A_HUMAN
|
||||||
| NC score | 0.003996 (rank : 87) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 64 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||