Please be patient as the page loads
|
SPY4_MOUSE
|
||||||
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |
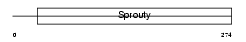
|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SPY4_MOUSE
|
||||||
| θ value | 5.58098e-151 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY4_HUMAN
|
||||||
| θ value | 3.61748e-150 (rank : 2) | NC score | 0.997043 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C004, Q6QIX2, Q9C003 | Gene names | SPRY4 | |||
|
Domain Architecture |
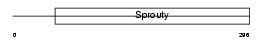
|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY3_HUMAN
|
||||||
| θ value | 4.39379e-55 (rank : 3) | NC score | 0.950792 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43610 | Gene names | SPRY3 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 3 (Spry-3). | |||||
|
SPY1_HUMAN
|
||||||
| θ value | 6.34462e-54 (rank : 4) | NC score | 0.935870 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY2_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 5) | NC score | 0.932076 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_HUMAN
|
||||||
| θ value | 2.75849e-49 (rank : 6) | NC score | 0.933341 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY1_MOUSE
|
||||||
| θ value | 4.70527e-49 (rank : 7) | NC score | 0.933154 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXV9 | Gene names | Spry1 | |||
|
Domain Architecture |
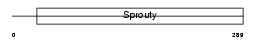
|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPRE2_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 8) | NC score | 0.498169 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE2_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 9) | NC score | 0.492806 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE1_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 10) | NC score | 0.497336 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE1_MOUSE
|
||||||
| θ value | 8.97725e-08 (rank : 11) | NC score | 0.495122 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q924S8, Q6PET8 | Gene names | Spred1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 12) | NC score | 0.510764 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE3_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 13) | NC score | 0.502247 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
ITB2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 14) | NC score | 0.056620 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
LRP1B_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.054060 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
KRA92_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.089254 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.053858 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
KR10B_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.092950 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.046693 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
DLK_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.045355 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.044902 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.048649 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.047703 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.046638 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.042440 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
NTCP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.022711 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12908, Q13839 | Gene names | SLC10A2, NTCP2 | |||
|
Domain Architecture |
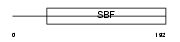
|
|||||
| Description | Ileal sodium/bile acid cotransporter (Ileal Na(+)/bile acid cotransporter) (Na(+)-dependent ileal bile acid transporter) (Ileal sodium-dependent bile acid transporter) (ISBT) (IBAT) (Apical sodium- dependent bile acid transporter) (ASBT) (Sodium/taurocholate cotransporting polypeptide, ileal) (Solute carrier family 10 member 2). | |||||
|
ZAN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.047498 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
ZAN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.049351 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.039985 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.040905 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.015693 (rank : 57) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.032219 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.040970 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
TSP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.040415 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35443, Q86TG2 | Gene names | THBS4, TSP4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.010396 (rank : 58) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.000856 (rank : 60) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.046867 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.049356 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.029966 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.037336 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
VEGFC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.025377 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97953, Q543R6 | Gene names | Vegfc | |||
|
Domain Architecture |
|
|||||
| Description | Vascular endothelial growth factor C precursor (VEGF-C) (Vascular endothelial growth factor-related protein) (VRP) (Flt4 ligand) (Flt4- L). | |||||
|
CRUM2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.039316 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
MYRIP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.019858 (rank : 55) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
ITB8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.035844 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
M3K14_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | -0.001160 (rank : 61) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||
|
MTR1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.001616 (rank : 59) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
MYRIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.017942 (rank : 56) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.063414 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.066049 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
EVL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.081040 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
EVL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.077391 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
KR107_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052330 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.055364 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.055816 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.064614 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA93_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.066987 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
KRA98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.063853 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.063446 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.054038 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
VASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.075893 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
VASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.079756 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
SPY4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.58098e-151 (rank : 1) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WTP2, Q9QXV7 | Gene names | Spry4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY4_HUMAN
|
||||||
| NC score | 0.997043 (rank : 2) | θ value | 3.61748e-150 (rank : 2) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C004, Q6QIX2, Q9C003 | Gene names | SPRY4 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 4 (Spry-4). | |||||
|
SPY3_HUMAN
|
||||||
| NC score | 0.950792 (rank : 3) | θ value | 4.39379e-55 (rank : 3) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43610 | Gene names | SPRY3 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 3 (Spry-3). | |||||
|
SPY1_HUMAN
|
||||||
| NC score | 0.935870 (rank : 4) | θ value | 6.34462e-54 (rank : 4) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O43609, Q6PNE0 | Gene names | SPRY1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY2_HUMAN
|
||||||
| NC score | 0.933341 (rank : 5) | θ value | 2.75849e-49 (rank : 6) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY1_MOUSE
|
||||||
| NC score | 0.933154 (rank : 6) | θ value | 4.70527e-49 (rank : 7) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QXV9 | Gene names | Spry1 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 1 (Spry-1). | |||||
|
SPY2_MOUSE
|
||||||
| NC score | 0.932076 (rank : 7) | θ value | 1.23823e-49 (rank : 5) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPRE3_HUMAN
|
||||||
| NC score | 0.510764 (rank : 8) | θ value | 2.61198e-07 (rank : 12) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2MJR0 | Gene names | SPRED3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE3_MOUSE
|
||||||
| NC score | 0.502247 (rank : 9) | θ value | 2.61198e-07 (rank : 13) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6P6N5, Q7TNJ8 | Gene names | Spred3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 3 (Spred-3). | |||||
|
SPRE2_HUMAN
|
||||||
| NC score | 0.498169 (rank : 10) | θ value | 2.36244e-08 (rank : 8) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z698, Q2NKX6 | Gene names | SPRED2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
SPRE1_HUMAN
|
||||||
| NC score | 0.497336 (rank : 11) | θ value | 8.97725e-08 (rank : 10) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE1_MOUSE
|
||||||
| NC score | 0.495122 (rank : 12) | θ value | 8.97725e-08 (rank : 11) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q924S8, Q6PET8 | Gene names | Spred1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SPRE2_MOUSE
|
||||||
| NC score | 0.492806 (rank : 13) | θ value | 5.26297e-08 (rank : 9) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q924S7 | Gene names | Spred2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 2 (Spred-2). | |||||
|
KR10B_HUMAN
|
||||||
| NC score | 0.092950 (rank : 14) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P60412 | Gene names | KRTAP10-11, KAP10.11, KAP18-11, KRTAP10.11, KRTAP18-11, KRTAP18.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-11 (Keratin-associated protein 10.11) (High sulfur keratin-associated protein 10.11) (Keratin-associated protein 18-11) (Keratin-associated protein 18.11). | |||||
|
KRA92_HUMAN
|
||||||
| NC score | 0.089254 (rank : 15) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ4 | Gene names | KRTAP9-2, KAP9.2, KRTAP9.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-2 (Keratin-associated protein 9.2) (Ultrahigh sulfur keratin-associated protein 9.2). | |||||
|
EVL_HUMAN
|
||||||
| NC score | 0.081040 (rank : 16) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UI08, O95884, Q7Z522, Q8TBV1, Q9UF25, Q9UIC2 | Gene names | EVL, RNB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
VASP_MOUSE
|
||||||
| NC score | 0.079756 (rank : 17) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70460, Q3TAP0, Q3TCD2, Q3U0C2, Q3UDF1, Q91VD2, Q9R214 | Gene names | Vasp | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.077391 (rank : 18) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
VASP_HUMAN
|
||||||
| NC score | 0.075893 (rank : 19) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P50552, Q6PIZ1, Q93035 | Gene names | VASP | |||
|
Domain Architecture |

|
|||||
| Description | Vasodilator-stimulated phosphoprotein (VASP). | |||||
|
KRA93_HUMAN
|
||||||
| NC score | 0.066987 (rank : 20) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ3 | Gene names | KRTAP9-3, KAP9.3, KRTAP9.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-3 (Keratin-associated protein 9.3) (Ultrahigh sulfur keratin-associated protein 9.3). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.066049 (rank : 21) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.064614 (rank : 22) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
KRA98_HUMAN
|
||||||
| NC score | 0.063853 (rank : 23) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BYQ0 | Gene names | KRTAP9-8, KAP9.8, KRTAP9.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-8 (Keratin-associated protein 9.8) (Ultrahigh sulfur keratin-associated protein 9.8). | |||||
|
KRA99_HUMAN
|
||||||
| NC score | 0.063446 (rank : 24) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BYP9 | Gene names | KRTAP9-9, KAP9.9, KRTAP9.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 9-9 (Keratin-associated protein 9.9) (Ultrahigh sulfur keratin-associated protein 9.9). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.063414 (rank : 25) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
ITB2_MOUSE
|
||||||
| NC score | 0.056620 (rank : 26) | θ value | 0.0563607 (rank : 14) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P11835, Q64482 | Gene names | Itgb2 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/P150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.055816 (rank : 27) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.055364 (rank : 28) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
LRP1B_MOUSE
|
||||||
| NC score | 0.054060 (rank : 29) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 606 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9JI18, Q8BZD3, Q8BZM7 | Gene names | Lrp1b, Lrpdit | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.054038 (rank : 30) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.053858 (rank : 31) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.052330 (rank : 32) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.049356 (rank : 33) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.049351 (rank : 34) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.048649 (rank : 35) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.047703 (rank : 36) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ZAN_HUMAN
|
||||||
| NC score | 0.047498 (rank : 37) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y493, O00218, Q96L85, Q96L86, Q96L87, Q96L88, Q96L89, Q96L90, Q9BXN9, Q9BZ83, Q9BZ84, Q9BZ85, Q9BZ86, Q9BZ87, Q9BZ88 | Gene names | ZAN | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.046867 (rank : 38) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.046693 (rank : 39) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.046638 (rank : 40) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
DLK_MOUSE
|
||||||
| NC score | 0.045355 (rank : 41) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q09163, Q07645, Q62208 | Gene names | Dlk1, Dlk, Pref1, Scp-1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (Preadipocyte factor 1) (Pref-1) (Adipocyte differentiation inhibitor protein) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.044902 (rank : 42) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.042440 (rank : 43) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.040970 (rank : 44) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.040905 (rank : 45) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
TSP4_HUMAN
|
||||||
| NC score | 0.040415 (rank : 46) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P35443, Q86TG2 | Gene names | THBS4, TSP4 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-4 precursor. | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.039985 (rank : 47) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
CRUM2_HUMAN
|
||||||
| NC score | 0.039316 (rank : 48) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5IJ48, Q5JS41, Q5JS43, Q6ZTA9, Q6ZWI6 | Gene names | CRB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 2 precursor (Crumbs-like protein 2). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.037336 (rank : 49) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
ITB8_HUMAN
|
||||||
| NC score | 0.035844 (rank : 50) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P26012 | Gene names | ITGB8 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-8 precursor. | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.032219 (rank : 51) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.029966 (rank : 52) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
VEGFC_MOUSE
|
||||||
| NC score | 0.025377 (rank : 53) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97953, Q543R6 | Gene names | Vegfc | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor C precursor (VEGF-C) (Vascular endothelial growth factor-related protein) (VRP) (Flt4 ligand) (Flt4- L). | |||||
|
NTCP2_HUMAN
|
||||||
| NC score | 0.022711 (rank : 54) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12908, Q13839 | Gene names | SLC10A2, NTCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ileal sodium/bile acid cotransporter (Ileal Na(+)/bile acid cotransporter) (Na(+)-dependent ileal bile acid transporter) (Ileal sodium-dependent bile acid transporter) (ISBT) (IBAT) (Apical sodium- dependent bile acid transporter) (ASBT) (Sodium/taurocholate cotransporting polypeptide, ileal) (Solute carrier family 10 member 2). | |||||
|
MYRIP_MOUSE
|
||||||
| NC score | 0.019858 (rank : 55) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3I4, Q8CFC0, Q8K4H5 | Gene names | Myrip, Slac2c | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
MYRIP_HUMAN
|
||||||
| NC score | 0.017942 (rank : 56) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFW9, Q8IUF5, Q9Y3V4 | Gene names | MYRIP, SLAC2C | |||
|
Domain Architecture |

|
|||||
| Description | Rab effector MyRIP (Myosin-VIIa- and Rab-interacting protein) (Exophilin-8) (Slp homolog lacking C2 domains c). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.015693 (rank : 57) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.010396 (rank : 58) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
MTR1L_HUMAN
|
||||||
| NC score | 0.001616 (rank : 59) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13585 | Gene names | GPR50 | |||
|
Domain Architecture |

|
|||||
| Description | Melatonin-related receptor (G protein-coupled receptor 50) (H9). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.000856 (rank : 60) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
M3K14_MOUSE
|
||||||
| NC score | -0.001160 (rank : 61) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 47 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUL6 | Gene names | Map3k14, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 14 (EC 2.7.11.25) (NF- kappa beta-inducing kinase) (Serine/threonine-protein kinase NIK). | |||||